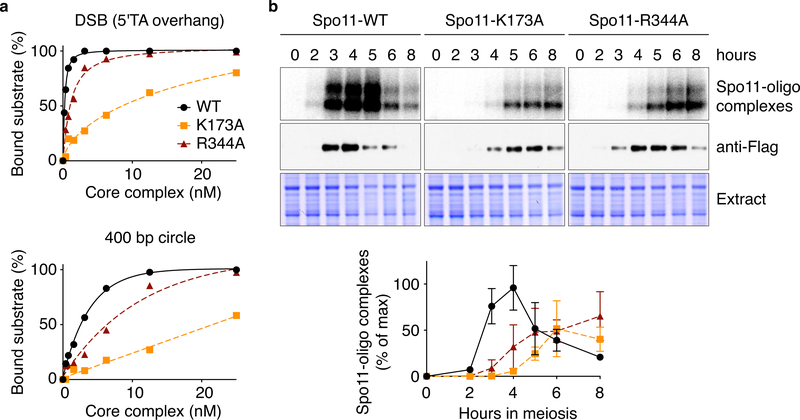
Fig. 6: Mutations that affect Spo11 DNA binding compromise DSB formation.
a. Binding of wild-type or mutant Spo11-containing core complexes to a 25-bp hairpin substrate with 5′-TA overhang (DSB) or a DNA mini-circle. The affinity of R344A was decreased 3-fold for DNA duplex and 4-fold for DNA ends. For K178A, the affinity was decreased 27-fold for the ends and could not be measured on duplex DNA because binding did not reach saturation.
b. Labeling of Spo11-oligo complexes. Representative gels of n = 2 assays are shown above; quantification is plotted below (mean and range from two experiments).
Uncropped blot and gel images and data for graphs are provided in the Source Data.