Fig. 1. Ribbon diagram of the structure of human SULT1E1-17β-estradiol (E2)-3’-phosphoadenosine 5-phosphate (PAP) complex showing the locations of amino acid residues involved in the SULT1E1 cSNPs studied.
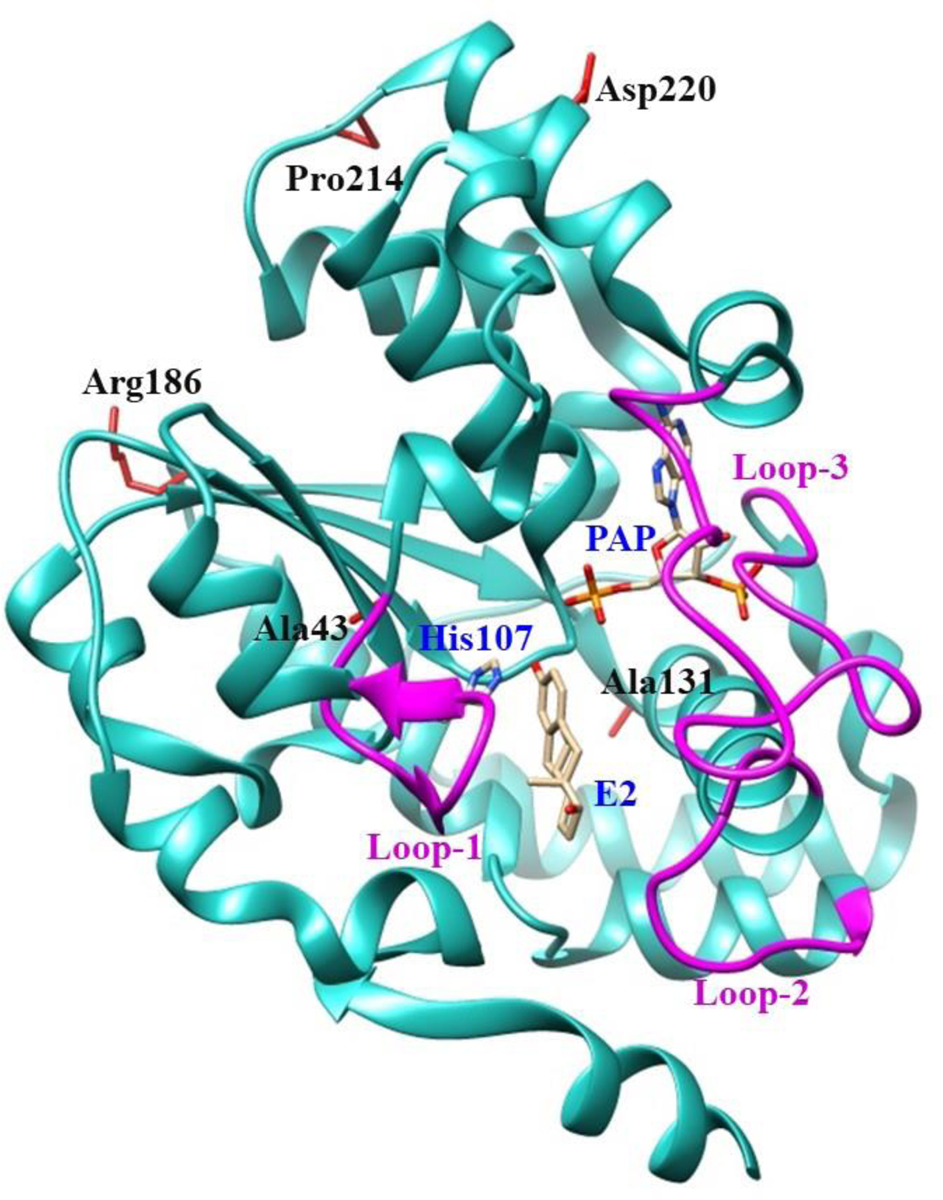
The structure of SULT1E1 (Protein Data Bank code: 4JVL), containing the polypeptide from Asp2 (N-terminus) to Glu293 (C-terminus), was depicted using Structure Comparison Analysis in USCF Chimera, a molecular modeling software [37]. E2 and PAP molecules in the structure are shown by bond structures. Loop-1 (Phe80-Asp90), loop-2 (Met144-Ser153), and loop-3 (Pro235-Gly25), shown by magenta color, form a gate for substrate entry [57, 58]. Side chains of the amino acid residues involved in the SULT1E1 cSNPs Ala43, Ala131, Arg186, Pro214 and Asp220 are indicated by bond structures (red color). PAP, (3′-phosphoadenosine 5′-phosphate); E2, (17β-Estradiol).
