Fig. 9. Hydrophobic interaction and hydrogen bond analyses of the SULT1E1 allozymes.
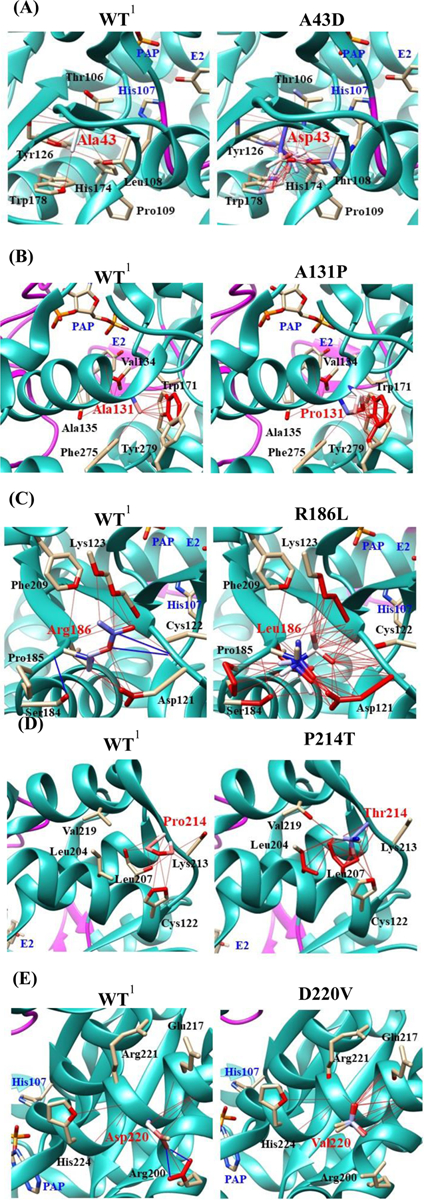
Atoms interacted with Ala43 (A), Ala131 (B), Arg186 (C), Pro214 (D), and Asp220 (E) are colored by the blue-white-red gradient (left panels; WT). Estimated interaction formed with Asp43 in A43D (A), Pro131 in A131P (B), Leu186 in R186L (C), Thr214 in P214T (D), and Val220 in D220V (E) are colored by the blue-white-red gradient (right panels; substituted). Side-chain conformation of a substituted residue was simulated using the Dunbrack backbone-dependent rotamer library [36]. Hydrophobic and hydrogen bond interactions of the substituted residues were also simulated by Find Clashes/Contacts tool in USCF Chimera software [37]. Top five-ranked rotamers of each substituted residue are modeled using the Dunbrack backbone-dependent rotamer library [36] and interaction was analyzed by Find Clashes/Contacts tool in USCF Chimera software. Hydrogen bonds formed with Cys122 (C) and Arg200 (E) are shown by blue slid lines (left panels). 1Wild-type human SULT1E1; PAP, (3’-phosphoadenosine 5’-phosphate); E2, (17β-Estradiol).
