Figure 6.
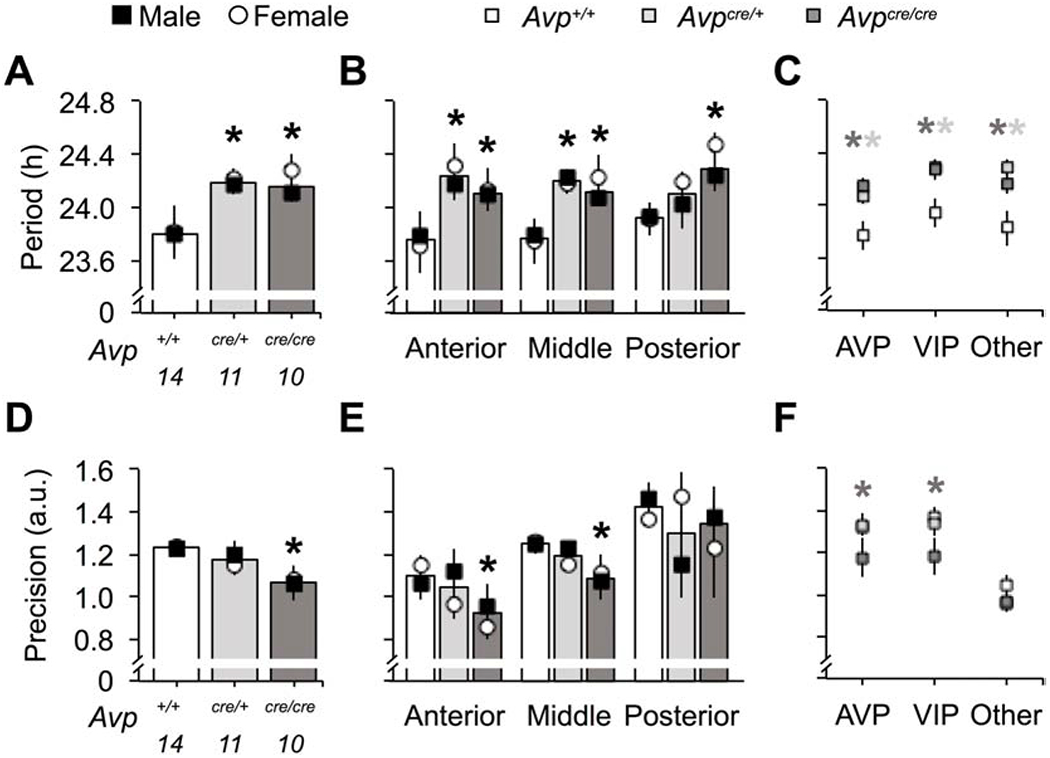
Avpcre/cre SCN neurons display increased cellular period and decreased cellular precision of PER2::LUC rhythms ex vivo. A-C. Cellular period of Avpcre/+ and Avpcre/cre SCN neurons is longer than wild-type (A, Genotype: F (2,34) = 6.3, p < 0.01, ηp2 = 0.3, Sex: F (1,34) = 0.56, p > 0.1, ηp2 = 0.02, Genotype*Sex: F (2,34) = 0.19, p > 0.1, ηp2 = 0.01) across the anteroposterior axis of the network (B, Genotype: F (2,104) = 10.9, p < 0.0001, ηp2 = 0.2, Sex: F (1,104) = 1.07, p > 0.1, ηp2 = 0.01, Slice: F (2,104) = 0.49, p > 0.1, ηp2 = 0.01, Genotype*Sex: F (2,104) = 0.24, p > 0.1, ηp2 = 0.006, Genotype*Slice: F (4,104) = 0.73, p > 0.1, ηp2 = 0.03, Sex*Slice: F (2,104) = 0.16, p > 0.1, ηp2 = 0.004, Genotype*Sex*Slice: F (4,104) = 0.18, p > 0.1, ηp2 = 0.008) and across different peptidergic regions (C, Genotype: F (2,104) = 13.8, p < 0.0001, ηp2 = 0.24, Peptide: F (2,104) = 1.42, p > 0.1, ηp2 = 0.03, Sex: F (1,104) = 1.29, p > 0.1, ηp2 = 0.01, Genotype*Peptide: F (4,104) = 0.34, p > 0.1, ηp2 = 0.02, Genotype*Sex: F (2,104) = 0.16, p > 0.1, ηp2 = 0.004, Peptide*Sex: F (2,104) = 0.57, p > 0.1, ηp2 = 0.01, Genotype*Sex*Peptide: F (4,104) = 0.45, p > 0.1, ηp2 = 0.02). D-F. Cellular precision is decreased in Avpcre/cre SCN neurons (D, Genotype: F (2,34) = 4.09, p < 0.05, ηp2 = 0.23, Sex: F (1,34) = 0.03, p > 0.1, ηp2 = 0.001, Genotype*Sex: F (2,34) = 0.23, p > 0.1, ηp2 = 0.02), with regional effects specific to the anterior and middle SCN (E, Genotype: F (2,104) = 3.68, p < 0.05, ηp2 = 0.08, Sex: F (1,104) = 0.06, p > 0.1, ηp2 = 0.0007, Slice: F (2,104) = 15.68, p < 0.0001, ηp2 = 0.27, Genotype*Sex: F (2,104) = 0.31, p > 0.1, ηp2 = 0.007, Genotype*Slice: F (4,104) = 0.18, p > 0.1, ηp2 = 0.008, Sex*Slice: F (2,104) = 0.24, p > 0.1, ηp2 = 0.006, Genotype*Sex*Slice: F (4,104) = 2.08, p = 0.09, ηp2 = 0.09) and to AVP- and VIP-expressing regions (F, Genotype: F (2,104) = 4.76, p < 0.05, ηp2 = 0.1, Peptide: F (2,104) = 20.91, p < 0.0001, ηp2 = 0.32, Sex: F (1,104) = 1.58, p > 0.1, ηp2 = 0.02, Genotype*Peptide: F (4,104) = 0.97, p > 0.1, ηp2 = 0.04, Genotype*Sex: F (2,104) = 1.12, p > 0.1, ηp2 = 0.03, Peptide*Sex: F (2,104) = 1.2, p > 0.1, ηp2 = 0.03, Genotype*Sex*Peptide: F (4,104) = 1.59, p > 0.1, ηp2 = 0.07). Location of peptidergic regions are illustrated in Figure 5. Numbers below abscissa represent sample size for each genotype, collapsed across sex (3-7/sex). a.u. = arbitrary units. Post hoc LSM Contrasts: * Differs from Avp+/+, genotype difference collapsed across sex, p < 0.05, (color coded for genotype in C and E). Other conventions as in Figure 1.
