Figure 1. A doxycycline-inducible CRISPRi system in S. pneumoniae.
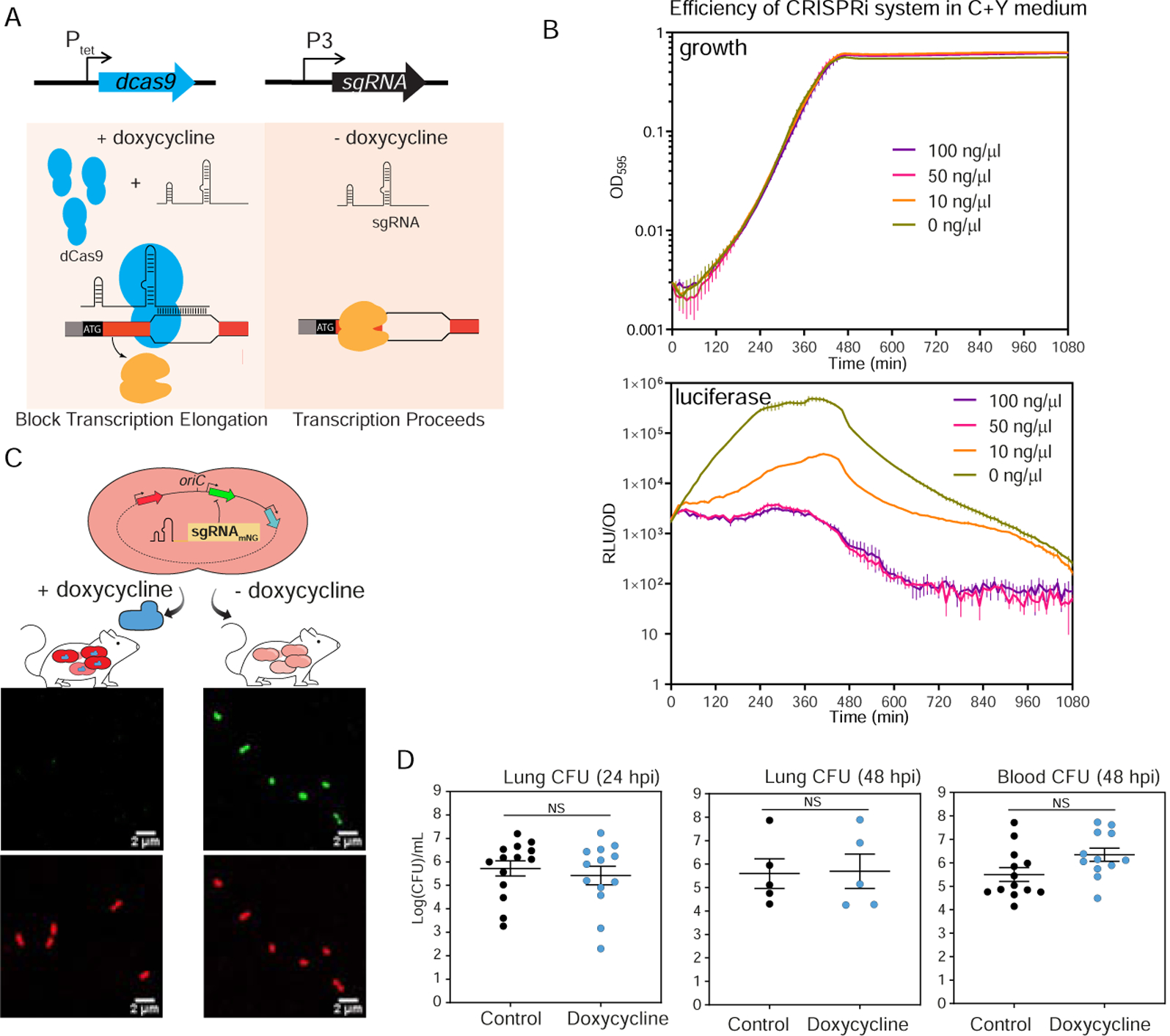
(A) The two key elements, dcas9 and sgRNA, were integrated into the chromosome of S. pneumoniae D39V and driven by a doxycycline-inducible promoter (Ptet) and a constitutive promoter (P3), respectively. With addition of doxycycline, dCas9 is expressed and binds to the target under the guidance of a constitutively expressed sgRNA. The specific dCas9-sgRNA binding to the target gene acts as a transcriptional roadblock. In the absence of the inducer, the target gene is transcribed. (B) The CRISPRi system was tested by targeting luc, which encodes firefly luciferase. The system was induced with doxycycline at different concentrations. Luciferase activity (RLU/OD) and cell density (OD595) were measured every 10 minutes. Top panel shows the growth and bottom panel shows the luciferase activity. The values represent averages of three replicates with SEM. (C) Reporter strain to assess in vivo activity of the doxycycline-inducible CRISPRi system. Strain VL2351 constitutively expresses mNeonGreen and mScarlet-I, and mNeonGreen is targeted by the sgRNA. Bacteria were collected from blood of mice on control or doxy-chow at 48 hpi and imaged with confocal microscopy in both the red and green channels. (D) Bacterial load at both lung and blood was quantified. Each dot represents a single mouse. Mean with SEM was plotted. There is no significant (NS) difference between the bacterial load in control- and doxycycline-treated mice (Mann Whitney U test).
