Fig. 1. Small molecule screen identifies antibiotics as suppressors of cell death in cellular models of human mitochondrial disease.
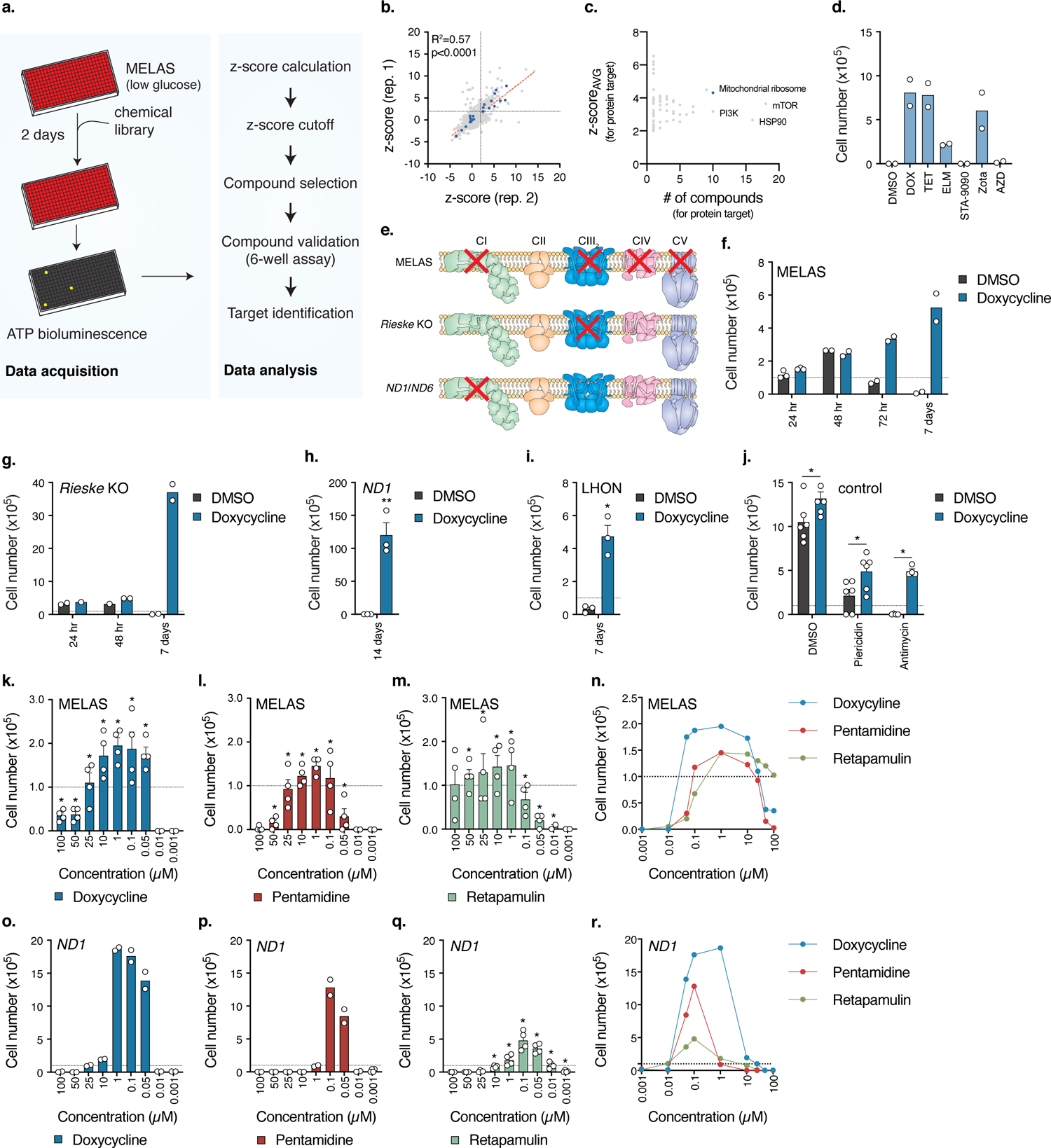
a. Schematic of the high-throughput small molecule screen to identify compounds that increase survival of MELAS cybrid cells. b. Plot of z-scores for each compound from two replicates. Antibiotics targeting the mitochondrial ribosome are highlighted in blue, R2 = 0.5704 (p<0.0001, Pearson). c. Plot of average z-scores for a particular protein against the number of compounds for that target. For compounds with multiple annotated target proteins, the compound z-score was incorporated into each protein target z-scoreAVG. d. 7 day low-glucose survival assay of MELAS cybrid cells treated with 3.3 μM compound (n=2 biologically independent samples). e. Comparison of electron transfer chain defects between the different cell lines used in the studies. MELAS, ND1, and ND6 (LHON) are human cybrid cell lines and Rieske is a mouse fibroblast cell line. Adapted from Rhee, H.W. et al.58 Reprinted with permission from AAAS. f–g. 7 day low-glucose survival assay of MELAS cybrid or Rieske cells treated with doxycycline (MELAS, n=2–3; Rieske, n=2 biologically independent samples). h. 14 day galactose survival assay of ND1 cybrid cells treated with doxycycline (n=3 biologically independent samples). i. 7 day galactose survival of LHON cybrid cells treated with doxycycline (n=3 biologically independent samples). j. Cell survival of U2OS control cells treated with 1 nM piericidin or 5 nM antimycin with 1 μM doxycycline in galactose media (n=2–3 biologically independent samples over n=2 independent experiments). k–m. 48 hour low-glucose survival assay of MELAS cybrid cells (n=2 biologically independent samples over n=2 independent experiments). n. Overlay of Fig. 1k–m. o–q. 7 day galactose survival assay of ND1 cybrid cells titrated with 3 structurally distinct antibiotics (Doxycycline, n=2; Pentamidine, n=2; Retapamulin, n=4 biologically independent samples). r. Overlay of Fig. 1o–q. Survival assays were initially seeded with 1.0×105 cells with media changed every 24 hours for low-glucose assays and 48 hours for galactose assays. Data are presented as mean values +/− s.e.m. error bars, Student’s t-test with a two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli, with Q = 5%, * q<0.05.
