Summary
Brain organoids closely recapitulate many features and characteristics of in vivo brain tissue. This technology in turn allows unprecedented possibilities to investigate brain development and function in the dish. Several brain organoid protocols have been established, and the studies have focused on validating the architecture, cellular composition, and function of the organoids. In future, the improved and advanced organoid models will enable us to understand cellular and molecular features of the developing brain. However, several obstacles, such as the quality of the organoids, 3D structural analysis, and measurement of the neural connectivity need to be improved. In this perspective, we will provide an overview of the current state of the art of the brain organoid field, with a focus on protocols and organoid characterization. Additionally, we will address the current limitations of this evolving field and provide an understanding of the current brain organoid landscape and insight toward the next steps.
Subject areas: neuroscience, tissue engineering
Graphical abstract

Neuroscience; tissue engineering
Introduction
Studies elucidating how the human brain develops and functions have been carried out over the past decades (Sousa et al., 2017). However, the lack of appropriate in vitro models, combined with inaccessibility to human brain tissue, has severely hindered research. Recent advances in brain organoid technology have provided opportunities to uncover the complex process of human brain development and function in vitro. In fact, brain organoids can faithfully recapitulate many structural, developmental, and functional aspects of the human brain. Therefore, this promising technology will provide insight into basic, developmental, and translational research.
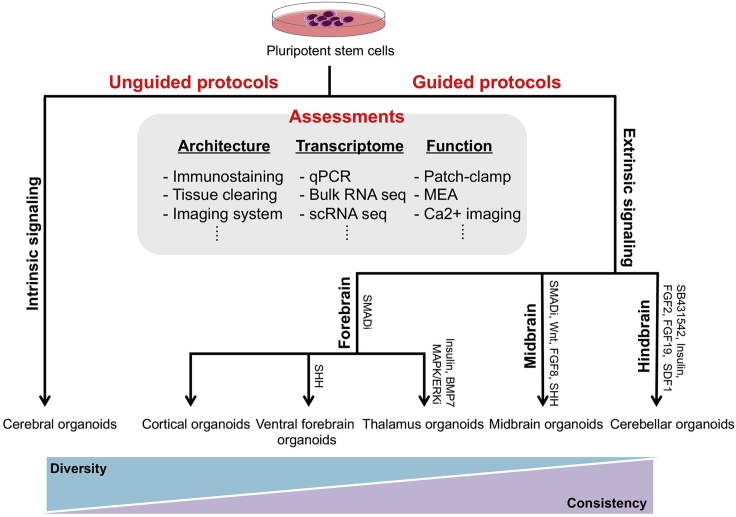
To date a variety of approaches have been described for the generation of brain organoids (Table 1). Methodologically, they can be divided into either unguided or guided protocols (Figure 1). Brain organoids produced through unguided protocols, the so-called cerebral organoids, are spontaneously differentiated from pluripotent stem cells (PSCs) without the inclusion of extrinsic factors, and the resulting organoids contain various brain regions. In contrast, in guided protocols, brain organoids are generated from PSCs through the addition of extrinsic factors to pattern to the desired regional identity. Both protocols can be adapted to reflect the critical aspects of human brain development. In addition, as organoid research progresses, so have the tools to analyze brain organoids at a higher resolution. For example, improved imaging techniques, multiple functional assays, and single-cell transcriptome analysis have all been applied to dissect brain organoids.
Table 1.
Overview of current brain organoid protocols
| Methodology | Regional identity | Type of organoid | Extrinsic factors | Assessment | Reference |
|---|---|---|---|---|---|
| Unguided method | Whole brain | Cerebral organoid | - | Immunostaining Calcium imaging |
(Lancaster et al., 2013) |
| Whole brain | Cerebral organoid | - | Immunostaining scRNA-seq MEA recording |
(Quadrato et al., 2017) | |
| Forebrain | Microfilament-engineered cerebral organoid | CHIR99021 (pulse) | Immunostaining Bulk RNA-seq Calcium imaging |
(Lancaster et al., 2017) | |
| Transplanted cerebral organoid | - | Immunostaining Calcium imaging |
(Mansour et al., 2018) | ||
| Air-liquid interface cerebral organoid | CHIR99021 (pulse) | Immunostaining scRNA-seq MEA recording Patch-clamp recording |
(Giandomenico et al., 2019) | ||
| Guided method | Cerebral cortex | Cortical tissue | Dkk-1, Lefty-A, and BMPRIA-Fc | Immunostaining | (Eiraku et al., 2008) |
| Cortical neuroepithelium | IWR1e and SB431542 | Immunostaining | (Kadoshima et al., 2013) | ||
| Cortical spheroid | Dorsomorphin and SB431542 | Immunostaining Microarray Calcium imaging Patch-clamp recording |
(Pasca et al., 2015) | ||
| Cortical organoid | Noggin and rhDkk1 | Immunostaining Bulk RNA-seq Patch-clamp recording |
(Mariani et al., 2015) | ||
| Forebrain organoid | Dorsomorphin, A83-01, WNT-3A, CHIR99021, and SB431542 | Immunostaining Bulk RNA-seq Calcium imaging Patch-clamp recording |
(Qian et al., 2016) | ||
| Cortical organoid | SB431542, LDN193189, and XAV939 | Immunostaining scRNA-seq Bulk RNA-seq Calcium imaging Patch-clamp recording |
(Xiang et al., 2017) | ||
| Cortical spheroid | Dorsomorphin and SB431542 | Immunostaining scRNA-seq Bulk RNA-seq Calcium imaging |
(Sloan et al., 2017) | ||
| Forebrain organoid | IWR1e and SB431542 | Immunostaining scRNA-seq |
(Velasco et al., 2019) | ||
| Ventral forebrain | Subpallium spheroid | Dorsomorphin, SB431542, aIWP2, and aSAG | Immunostaining scRNA-seq Calcium imaging Patch-clamp recording |
(Birey et al., 2017) | |
| Ventral organoid | aIWP2 and aSAG | Immunostaining | (Bagley et al., 2017) | ||
| Ventral forebrain organoid | Noggin, SB431542, CHIR99021, aSHH, and aPurmorphamine | Immunostaining Calcium imaging |
(Kim et al., 2019) | ||
| Medial ganglionic eminence | MGE organoid | SB431542, LDN193189, and XAV939, aSHH, and aPurmorphamine | Immunostaining scRNA-seq Bulk RNA-seq Calcium imaging Patch-clamp recording |
(Xiang et al., 2017) | |
| Thalamus | Thalamus organoid | SB431542, LDN193189, Insulin, aPD0325901, and aBMP7 | Immunostaining scRNA-seq Calcium imaging Patch-clamp recording |
(Xiang et al., 2019) | |
| Hypothalamus | Hypothalamus organoid | SB431542, LDN193189, aWNT3A, aSHH, and aPurmorphamine | Immunostaining | (Qian et al., 2016) | |
| Midbrain | Midbrain organoid | SB431542, Noggin, CHIR99021, aSHH, and aFGF8 | Immunostaining Bulk RNA-seq Patch-clamp recording |
(Jo et al., 2016) | |
| Midbrain organoid | SB431542, LDN193189, CHIR99021, aFGF8, aSHH, and aPurmorphamine | Immunostaining | (Qian et al., 2016) | ||
| Cerebellum | Cerebellar tissue | SB431542, Insulin, FGF2, aFGF19, and aSDF1 | Immunostaining Patch-clamp recording |
(Muguruma et al., 2015) | |
| Hippocampus | Hippocampal tissue | SB431542, IWR1e, aCHIR99021, and aBMP4 (pulse) | Immunostaining Calcium imaging Patch-clamp recording |
(Sakaguchi et al., 2015) | |
| Choroid plexus | Choroid plexus tissue | SB431542, IWR1e, aCHIR99021, and aBMP4 | Immunostaining | (Sakaguchi et al., 2015) | |
| Choroid plexus organoid | aBMP4 and aCHIR99021 (pulse) | Immunostaining scRNA-seq Mass spectrometry |
(Pellegrini et al., 2020) | ||
| Spinal cord | Neuromuscular organoid | aCHIR99021, aFGF2, HGF, and IGH | Immunostaining scRNA-seq Calcium imaging MEA recording |
(Faustino Martins et al., 2020) |
Patterning factors.
Figure 1.
Methods to generate and analyze brain organoids
Unguided methods that do not use specific chemicals or inhibitors produce cerebral organoids with a high cellular diversity but with higher heterogeneity. Guided methods use signaling activators or inhibitors to produce region-specific brain organoids. Architecture, cellular diversity, and function of the brain organoids are assayed by multiple methods.
In this perspective, we will provide an overview of current brain organoid technology and address some of the most critical questions in the field. Specifically, we will discuss current methods in generating region-specific organoids and the integration of cells from non-neuroectodermal lineages. We will highlight the limitations and future perspectives of brain organoid technology. We anticipate that brain organoids will open up new avenues allowing a deeper understanding of both normal and pathogenic human brain development and function.
Current state-of-the-art brain organoid generation
Differentiation of human PSCs (hPSCs) into brain organoids is performed in a step-wise manner, composed of neuroectodermal induction, followed by patterning of regional identity, and finally neural maturation. The addition of specific growth factors or inhibitors or their omission during neuroectoderm induction determines cell fate at the end of differentiation (Figure 1). Broadly, unguided methods refer to neuroectodermal induction without the addition of extrinsic factors, whereas guided methods utilize small molecules and/or growth factors to produce brain organoids with a specific regional identity (Figure 2). The first described protocols developed by Lancaster and colleagues to generate cerebral organoids utilized an unguided approach (Lancaster and Knoblich, 2014; Lancaster et al., 2013). In this protocol, cerebral organoids are self-organized and spontaneously differentiated from hPSCs without extrinsic factors and in the presence of the extracellular matrix (ECM) Matrigel. The ECM functions to support three-dimensional (3D) structure and neuroepithelial expansion. Remarkably, the resulting organoids displayed an array of regional identities of the brain. Cellular and regional diversity of cerebral organoids provide a unique opportunity to investigate their interaction in whole-brain development. On the other hand, this diversity is a potential limitation that has to be addressed when performing quantitative studies in cerebral organoids. To address this, Lancaster and colleagues have made efforts to standardize the workflow by applying poly(lactide-co-glycolide) copolymer fiber microfilaments when generating elongated embryoid bodies (EBs) (Lancaster et al., 2017). This increased the surface area, improving the reproducibility of neural induction and subsequent cortical development.
Figure 2.
Patterning and morphogens in the neural tube
In the fetal brain, the dorsoventral and anteroposterior (rostrocaudal) developmental fate of the primary vesicles are determined by the morphogens. Following the default neuroectoderm commitment by inhibition of BMP signaling, the WNT and SHH signals determine the dorsoventral axis, and FGF8 signal determines the rostrocaudal fate. The combination of the agonists and antagonists of these signaling pathways is used to generate the regionally defined brain organoids in Figure 1.
In contrast, guided approaches utilize extrinsic signaling factors to choreograph the differentiation to produce region-specific brain organoids. The Sasai group reported one of the first descriptions of regionalized brain organoids, a combination of floating EB-like aggregates (SFEBq) under serum-free conditions supplemented with bone morphogenetic protein (BMP), transforming growth factor (TGF)-beta, and Wnt antagonists to guide the production of cortical tissue (Eiraku et al., 2008). The resulting tissue displayed polarized cortical neuroepithelia, along with a distinctly organized ventricular zone (VZ) and cortical plate (CP). The inhibition of BMP and TGF-beta signaling has been broadly applied to make efficient neuroectoderm induction and to produce cortical neurons (Chambers et al., 2009; Kim et al., 2010). Several groups have employed dual SMAD and Wnt inhibition to coax a dorsal forebrain identity during brain organoid generation, namely, cortical organoids (Kadoshima et al., 2013; Mariani et al., 2015; Pasca et al., 2015; Qian et al., 2016; Xiang et al., 2017). These cortical organoids recapitulate the features of cortical development such as temporal order of neurogenesis and functional cortical neurons. Additionally, the Ming group developed a miniaturized spinning bioreactor to reduce cost but importantly to minimize heterogeneity and the variability of derived cortical organoids (Qian et al., 2016). These organoids produced a well-developed outer subventricular zone (oSVZ) and diversity of neuronal subtypes. Recently, oSVZ containing an abundant population of outer radial glia cells was markedly expanded through STAT3 activation in cortical organoids (Watanabe et al., 2017). Thus, it enables us to study human-specific features of brain development.
During forebrain development, dorsal-ventral identity is determined by spatial and temporal signals. An important player is sonic hedgehog (SHH) signaling, which is critical in ventral telencephalic patterning (Figure 2) (Fuccillo et al., 2004). To specify brain organoids with either a ventral forebrain or medial ganglionic eminence (MGE) domain, neuroectoderm-committed brain organoids, generated through dual SMAD inhibition, were exposed to SHH and its agonist purmorphamine (Bagley et al., 2017; Birey et al., 2017; Xiang et al., 2017). The ventral forebrain organoids contained various GABAergic interneuron subtypes with physiological properties, whereas MGE organoids mostly displayed somatostatin (SST)+ interneurons, exclusively produced from the MGE. Notably, these specific forebrain organoids have allowed the modeling of tangential migration of interneurons into cortex by fusing dorsal forebrain organoids with ventral forebrain organoids (Bagley et al., 2017; Birey et al., 2017; Kim et al., 2019; Xiang et al., 2017). In turn, applying this novel strategy will allow researchers to explore the interaction of the developing dorsal and ventral cortical regions.
Although most studies have focused on brain organoids representing the forebrain of telencephalic origin, a number of other region-specific brain organoids have been described leveraging off the ability to drive region-specific patterning as described above. For example, protocols have been reported for organoids with a diencephalic identity (Figure 2). The diencephalon is a part of the vertebrate neural tube that gives rise to the thalamus and hypothalamus. To generate hypothalamic organoids, the Ming group used a combination of growth factors (WNT3A and SHH) and the Smoothened (Smo) receptor agonist, purmorphamine, resulting in organoids that contained neuropeptidergic hypothalamic neurons (Qian et al., 2016). Our laboratory described an approach to produce thalamic organoids, by pre-patterning with insulin and the MAPK/ERK inhibitor PD0325901. The role of PD0325901 was to suppress excess insulin-induced caudalization to a mesencephalon fate. Finally, BMP7 was used to drive a diencephalic fate toward a thalamic identity (Xiang et al., 2019). Indeed, thalamus and cortex interactions are highly involved in sensory processing and cognitive functions (Lopez-Bendito and Molnar, 2003; Ouhaz et al., 2018). Nevertheless, currently available in vitro human models have not yet been developed. Therefore, thalamus organoids will be critical to understand thalamic development and interaction between both thalamus and cortex.
Patterning to the mid- and hindbrain regions of the brain have also been an area of intense research. The mesencephalon (midbrain) gives rise to three main areas, the colliculi, the tegmentum, and the cerebral peduncle, whereas the rhombencephalon (hindbrain) gives rise to the pons, cerebellum, and medulla oblongata. Their patterning and morphogenesis are highly regulated by the midbrain-hindbrain boundary (Gibbs et al., 2017).
The generation of midbrain organoids was reported by Jo et al. (2016). They utilized EBs, which were subjected to dual SMAD inhibition in combination with the WNT agonist CHIR99021, to pattern to mesencephalon. These were exposed to a SHH agonist along with FGF8 for floor plate induction (Figure 2). The resulting midbrain organoids contained functional dopaminergic neurons and produce neuromelanin-like granules, similar to those found in the substantia nigra domain. Another report from the Song lab used a very similar approach as mentioned earlier, but after the derivation of floor plate they were transferred to a miniaturized spinning bioreactor, producing mid-brain organoids that contained TH-positive dopaminergic neurons (Qian et al., 2016). To generate the most caudal region of brain, dual SMAD inhibition was employed in combination with FGF2 and insulin treatment. This was sufficient to produce polarized cerebellar tissue, generating cerebellar plate neuroepithelium (Muguruma et al., 2015). Additional treatment of FGF19 and SDF1 promoted a dorsoventral polarity and rhombic-lip-like structure in the 3D cerebellar tissues (Muguruma et al., 2015).
The cortical hem is a dorsal midline structure that provides a source of WNT and BMP signaling in the dorsomedial telencephalon, playing a key role in formation of the dorsomedial telencephalic tissues such as the hippocampus and choroid plexus (Lun et al., 2015). Following this developmental principle, the Sasai group applied a transient WNT (through the agonist CHIR99021) and BMP4 treatment to neuroectoderm to recapitulate hippocampal primordium tissues, whereas a prolonged activation of WNT and BMP produced choroid plexus-like tissue (Sakaguchi et al., 2015). However, this study did not investigate the functional attributes of the choroid plexus such as production or secretion of cerebrospinal fluid (CSF). In recent studies, structurally and functionally well-defined choroid plexus organoids have been generated (Pellegrini et al., 2020). Similar to the Sasai report, BMP4 and CHIR99021 were used to generate a choroid plexus-like identity during cerebral organoid generation. Importantly, choroid plexus organoids formed a tight barrier and produced a fluid resembling CSF. These organoids will enable investigation of the function of the choroid plexus during brain development and provide an important tool to assess the permeability of new drugs.
Importantly, one must consider the most suitable brain organoid method for a given study. For example, cerebral organoids generated through unguided protocols were applied to the study of neurodevelopmental disorder displaying dramatic phenotypes such as microcephaly (Lancaster et al., 2013). In contrast, due to high degree of consistency, the SFEBq-based method developed by the Sasai group (guided protocols) was used for high-throughput screening of brain organoids (Durens et al., 2020). In general, the unguided protocols are suited to investigate overt phenotypes of neurodevelopmental disorder such as microcephaly, macrocephaly, and periventricular heterotopia (PH) (Klaus et al., 2019; Lancaster et al., 2013; Li et al., 2017; O'Neill et al., 2018). Unguided protocols are also useful for studying communication between brain regions because cerebral organoids display multiple regional identities within a single organoid (Quadrato et al., 2017; Renner et al., 2017). Conversely, cortical organoids produced through guided protocols exhibit reproducible brain identities with relatively homogeneous morphology (Pasca et al., 2015; Qian et al., 2016; Velasco et al., 2019; Xiang et al., 2017). Guided protocols enable researchers to perform high-throughput screening using brain organoids. Thus, this approach is more suited to quantitative analysis. Furthermore, brain region-specific organoids generated by guided protocols can be fused into assembloids (Bagley et al., 2017; Xiang et al., 2017, 2019). These methods can be used for modeling neurodevelopmental disorders exhibiting disruption of cell migration and neural circuit such as Timothy syndrome (Birey et al., 2017).
Furthermore, when deciding which protocol to utilize for generating brain organoids, technical issues should be considered. In general, the process of brain organoid culture takes several months. Thus, a slight difference at the beginning of culturing will make a huge difference at the end of culturing. It is necessary to generate EBs with uniform size and morphology at the beginning that will minimize the heterogeneity of brain organoids during organoid generation (Guo et al., 2020). Moreover, to avoid resource loss, a more accurate prediction system for differentiation potential that is not simple morphology-based should be developed in the future. In addition, complicated manual steps are needed during brain organoid generation such as transferring manual organoids (Lancaster and Knoblich, 2014; Xiang et al., 2018). In particular, the step of embedding cerebral organoids into Matrigel is essential in unguided protocols (Lancaster and Knoblich, 2014; Lancaster et al., 2013). This is the greatest technical challenge during cerebral organoid generation. However, this laborious step is not necessary in guided protocols.
In fact, brain organoid methods require special equipment and culture plates. For instance, orbital shakers or spinning bioreactors have been widely used at the later stage of culture for enhancing oxygen exchange in both unguided and guided protocols (Lancaster et al., 2013; Qian et al., 2016; Xiang et al., 2017). Moreover, brain organoids should be cultured in low-attachment plates for suspension culture in the presence of a variety of small molecules and/or growth factors. Hence, both unguided and guided protocols typically come at a high financial cost for brain organoid culture. In general, one of the major costs in unguided protocols is the use of Matrigel, whereas a variety of high-cost small molecules are needed for patterning in guided protocols. In addition to small molecules, dissolved Matrigel was added during organoid maturation in some guided protocols (Kadoshima et al., 2013; Qian et al., 2016). Therefore, guided protocols have a slightly higher cost than unguided protocols due to the necessity of using additional factors.
In short, unguided and guided protocols have distinct limitations and a multitude of benefits. Unguided protocols are suitable for modeling overt phenotypes and studying brain region interaction within a single organoid, whereas guided protocols can be used for high-throughput screening. Moreover, fusion of brain region-specific organoids enables researchers to model cell migration and neural circuit between brain regions. Compared with unguided protocols, guided protocols are much less laborious but higher cost. When deciding which protocol to use for generating brain organoids, there are several considerations such as the scope of study, technical challenges, and costs. These considerations will assist researchers in choosing the best protocol for successfully implementing a brain organoid experiment.
Molecular and neurophysiological techniques for analyzing brain organoids
Brain organoids have a complex organization resembling the developing brain. For example, like the cortex, which is composed of six laminar layers, cortical organoids display distinct layers. The cortical neurons arising from VZ make radial migration and accumulate in the CP in an inside-out manner (Bystron et al., 2008). This is also well reproduced in cortical organoids. However, the complexity observed within brain organoids provides new challenges with regard to their analysis. This is in terms of cellular structural analysis, molecular analysis (to define the cellular identities), and neurophysiological analysis to define the function of neurons, which are a basic necessity (Figure 1).
To characterize the structural organization of organoids, conventional immunostaining against markers on sectioned organoids have been mainly employed. In fact, brain organoids contain VZs harboring proliferating apical progenitors expressing radial glial markers (SOX2 and PAX6). TBR2+ Intermediate progenitor cells are found in the subventricular zone (SVZ) (Kadoshima et al., 2013; Lancaster et al., 2013; Pasca et al., 2015; Qian et al., 2016; Xiang et al., 2017). Importantly, cells expressing HOPX and PTPRZ1, markers for human outer radial glia, are observed in the oSVZ, which are not found in rodent cortex (Qian et al., 2016). Moreover, brain organoids exhibit different cortical layers expressing specific markers. In the early stage, the population of deep layer neurons expressing TBR1 and CTIP2 is increased, whereas SATB2+ and CUX1+ upper layer neurons are found later in organoids. However, most brain organoids show restricted cortical layers (Kadoshima et al., 2013; Lancaster et al., 2013; Pasca et al., 2015; Xiang et al., 2017). Furthermore, MAP2+ mature neurons as well as GFAP+ astrocytes and OLIG2+ oligodendrocytes are observed at later stages of brain organoids (Madhavan et al., 2018; Marton et al., 2019; Pasca et al., 2015; Sloan et al., 2017). Thus, these organized structure and cellular diversity of brain organoids should be validated by using multiple cell-type-specific antibodies. However, sections of the organoids under analysis are randomly selected, and without the spatial organization, could lead to a bias in analysis. Random selection in field of analysis is simple and accurate particularly in large sample population. However, the selected samples do not represent the whole organoids, and thus whole-mount-staining and objective analysis will be needed. In addition, quantification of immunostaining images is also challenging. Detection methods and artifacts from non-specific binding of antibody should be considered for reliable quantification (Yaziji and Barry, 2006). To preserve the 3D spatial information within the organoid, whole organoids can be optically cleared and imaged at high resolution in 3D (Dekkers et al., 2019). This technique can provide a better understanding of the cellular complexity, cell types present, cell shape, and intracellular processes within the organoid in question. However, the accessibility of antibodies for analysis and the identification of the given structures are still challenging, especially in brain organoids where distinct neurons play different roles. In addition, the axonal and dendritic connectivity need to be visualized and defined.
Another area that has been rapidly evolving to resolve cellular heterogeneity within organoids is single-cell-based genomics approaches (Polioudakis et al., 2019; Poulin et al., 2016; Zeisel et al., 2015). In particular, single-cell RNA sequencing (scRNA-seq) has revealed cellular identity within complex tissues at the single-cell level (Tang et al., 2009). One of earliest scRNA-seq studies compared cerebral organoids against human fetal neocortex, demonstrating that the cell types, neuronal lineage relationships, and gene expression profiles were comparable (Camp et al., 2015). Recent advances in scRNA-seq have provided a large-scale experiment to compare gene expression between brain organoids and primary human tissues (Amiri et al., 2018; Bhaduri et al., 2020; Pollen et al., 2019; Velasco et al., 2019). Furthermore, droplet-based microfluidics platforms, such as the 10X Genomics platform (Chromium), are highly efficient and provide robust throughput with remarkably reduced cost. As a consequence of robustness, throughput, and cost, scRNA-seq is now widely used throughout the brain organoid field (Birey et al., 2017; Camp et al., 2015; Giandomenico et al., 2019; Pellegrini et al., 2020; Quadrato et al., 2017; Velasco et al., 2019; Xiang et al., 2017, 2019).
An example of high-throughput droplet-based scRNA-seq application is by Velasco and colleagues, who compared four different types of brain organoids (whole-brain organoids, dorsal organoids, dorsal spheroids, ventral spheroids) (Velasco et al., 2019). The transcriptional profiles of over 160,000 cells isolated from 21 individual organoids were used. The outcome of this study suggested that dorsal forebrain organoids displayed a similar cellular diversity as human cerebral cortex with high reproducibility. More recently, systematic approaches to compare scRNA-seq datasets from brain organoids generated by various protocols were performed (Tanaka et al., 2020). This study identified both unique differentiation trajectories and transcriptional biases emerging from the different brain organoid protocols when compared with the fetal brain. Indeed, brain organoids contain different subtypes of neurons as well as glial cells such as astrocytes and oligodendrocytes (Madhavan et al., 2018; Marton et al., 2019; Pasca et al., 2015; Sloan et al., 2017). Moreover, protocol differences can bias differentiation of cell types that are not typically present otherwise such as oligodendrocyte spheroids (Madhavan et al., 2018; Marton et al., 2019). Hence, during the analysis of scRNA-seq data, determination of cell identities through annotation is the most important step for revealing cellular identities in brain organoids. However, in general, annotation is manually performed based on the expression of marker genes (Muraro et al., 2016). This step is time-consuming and heavily dependent on prior knowledge, leading to inconsistent results. Recently, to improve the efficiency and reproducibility of annotation step, various automatic tools for annotating cells have been developed (Cao et al., 2020; Ekiz et al., 2020; Shao et al., 2020). These will enable researchers to annotate the cells at a fully automated mode.
Functional characterization, especially the electrophysiological properties of neurons, is critical to define the degree of maturation and the identity of neurons within organoids (Tambalo and Lodato, 2020). However, the time needed to establish neuronal networks should be considered. Indeed, neuronal activity was not found in 4-month-old organoids, whereas 8-month old organoids were functional in terms of spontaneous activity and excitatory monosynaptic connections (Quadrato et al., 2017). Moreover, it is elusive how brain organoids are functionally mature. Thus, this should be addressed further to make it clear whether alteration of their functionality is originated from the functional phenotype of disease or an artifact of long-term culture. In fact, at the later stages, brain organoids have the features of electrophysiological maturation as evidenced by complex network bursting and synchronized burst firings, which are distinct characteristics of functional neural networks and synaptic signal propagation. Immature brain organoids displayed immature neuronal activities such as random and low amplitude (Fair et al., 2020). Therefore, these electrophysiological features can be used as the criteria for maturation of brain organoids. Currently, the electrophysiological function of brain organoids are validated by various experimental approaches (Poli et al., 2019).
Using the patch-clamp approach, several studies have demonstrated that neurons in cerebral organoids and cortical organoids display both action potentials and spontaneous inhibitory/excitatory postsynaptic potentials (Birey et al., 2017; Giandomenico et al., 2019; Mariani et al., 2015; Pasca et al., 2015; Qian et al., 2016; Xiang et al., 2017, 2019). Furthermore, sensitivity to antagonists of glutamate and GABA receptors provided definitive evidence for the identity of functional excitatory or inhibitory neurons. It is well acknowledged that the neural maturation is a long-term process, and this is confirmed by older organoids tending to carry more mature neurons (Pasca et al., 2015; Renner et al., 2017; Trujillo et al., 2019). Electrophysiological recording is performed on randomly chosen cells in slices sectioned from organoids, and the action of sectioning leads to loss of 3D structures within the organoid. In addition, patch clamp is laborious and cannot determine the interconnectivity present in the brain organoid. Thus, the microelectrode arrays (MEAs) approach has been used as a functional assay in brain organoid studies (Giandomenico et al., 2019; Trujillo et al., 2019). While patch-clamping provides information on the intracellular electrophysiological features of the cell, MEA approach provides information via extracellular recording. An advantage of this method is the ability to monitor large number of neurons at multiple sites simultaneously. In addition, calcium imaging based on Ca2+ probes or engineered Ca2+ reporters, such as GCaMP6, have been widely applied to analyze the neural activity of brain organoids. Ca2+ imaging in neurons is a direct indicator of neural activity, because calcium ions are released from intraneural storage, in mature neurons, spontaneously as well as when stimulated, thus representing neural activity (Grienberger and Konnerth, 2012). Several studies have demonstrated spontaneous calcium surges in brain organoids; this indicates that neuronal networks have been established (Birey et al., 2017; Kim et al., 2019; Lancaster et al., 2013, 2017; Mansour et al., 2018; Pasca et al., 2015; Qian et al., 2016; Xiang et al., 2017, 2019). However, there is an urgent need for more advanced techniques to characterize brain organoids both anatomically and physiologically.
Limitation of brain organoid technology
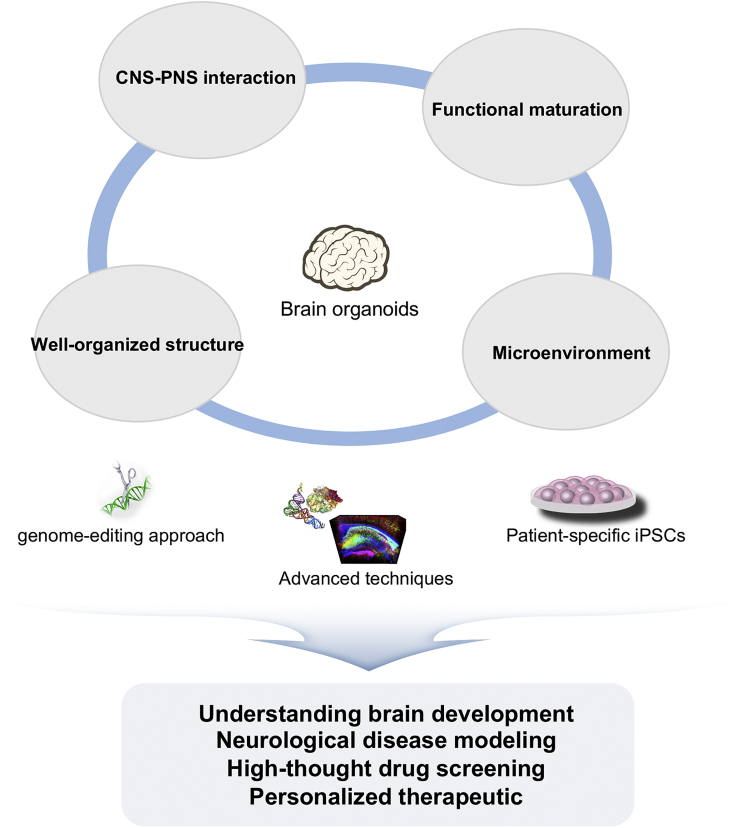
The advantages in utilizing brain organoid technology have been outlined, but as with any emerging technologies there are some limitations (Figure 3). One of major issues is that the majority of brain organoids, even at long-term culture, resemble a second-trimester fetal brain, in terms of structure, function, and transcriptome. Although these organoids are useful in probing the early neuroectoderm commitment and cortical development, they may not be suitable to investigate the physiological aspects of adult brain and to model late-onset neurodegenerative diseases. Human neurons intrinsically require long-term maturation (Zhao et al., 2008); the suboptimal differentiation conditions employed in vitro, such as a lack of microenvironment, are probably responsible for the observed immature neural states. Indeed, the pattern of local field potential in 10-month-old cortical organoids resembled electrophysiological signatures in human preterm neonatal electroencephalography (EEG). However, the complex and continuous pattern of network activity observed in adult EEG was not displayed (Trujillo et al., 2019). Moreover, maintaining the cortical organoids in a healthy condition is challenging during organoid culture.
Figure 3.
Future perspectives in brain organoids
Developing brain organoids with structurally better organized, reproducing the CNS-PNS connection, and physiological environment is needed. The improved brain organoids system will be critical to model human brain development and brain disorders, and to perform drug discovery and personalized therapeutics.
During in vivo brain development, the blood vessels penetrating from the pia mater play a crucial role in both scaffolding and blood flow during brain development (Kojima et al., 2010). In addition, neural differentiation and maturation cannot occur without sufficient oxygen and nutrient supply. However, brain organoids do not contain functional vasculature, and here may lie the problem. To address this, a number of groups have attempted to address the issue of vascularization in brain organoids. A direct approach was to co-culture brain organoids with endothelial cells (Pham et al., 2018; Shi et al., 2020). The organoids contained a well-developed vascular system and exhibited an improved maturity. However, neither capillaries nor the functionality of the vasculature were fully addressed. An approach was employed by the Gage lab, where they transplanted brain organoids into adult mouse brains (Mansour et al., 2018). The resulting engrafted brain organoids displayed progressive neural differentiation and maturation, gliogenesis, and unprecedented axonal outgrowth. Importantly, a functional vasculature system within the organoid was confirmed by active blood flow through the vessels. More recently, genetically engineered brain organoids have been developed (Cakir et al., 2019). The organoids contained ETV2 engineered cells that could be controlled in an inducible manner. Notably, the ETV2-expressing population within organoids contributed to vascularization, leading to enhanced functional maturation with blood-brain barrier characteristics. These vascularized brain organoids can more precisely recapitulate the physiological features of the brain. However, it still remains elusive whether vasculature in brain organoids can deliver adequate oxygen and nutrients alone.
A feature of the brain is cavities, known as ventricles, which contain CSF produced by choroid plexus’ activity. These ventricles are surrounded by an ependymal layer called the VZ at the embryonic stage. In the postnatal stage, SVZ lines the ventricles. During neurogenesis, cortical neurons migrating from VZ form radially organized structures, called the CP, consisting of six distinct layers of neurons. However, brain organoids exhibit limited spatial organization and complexity compared with the developing fetal brain. In general, the six cortical layers are not clearly separated in brain organoids. It is impossible to discriminate each layer in the organoids based on layer-specific neuron distribution and morphology. A miniaturized spinning bioreactor seems to produce more diverse neuronal subtypes of all six cortical layers despite rudimentary separation (Qian et al., 2016). Although brain organoids display limited organized structure, defined neuronal migration processes are observed (Bagley et al., 2017; Xiang et al., 2017). Neuronal migration defects have also been investigated using brain organoids in various diseases such as PH (Birey et al., 2017; Klaus et al., 2019). Moreover, the majority of studies show that brain organoids contain lumen, resembling a ventricle, with surrounding neural progenitor cells. However, control of ventricle-like structure in the size and number has been very challenging. In the cortex, there are only two cortical ventricles as opposed to multiple ventricle-like structures observed in brain organoids. A recent study demonstrated that elongating EBs with microfilaments could coax more consistent enlarged ventricle-like lumen with neuroepithelium during organoid generation (Lancaster et al., 2017). Recently, a protocol for generating choroid plexus organoids secreting CSF was established (Pellegrini et al., 2020). In this study, choroid plexus organoids displayed lateral ventricle identity based on transcriptomic signatures. These organoids would serve as excellent models to investigate the ventricular system in vitro. More such studies are required to fully understand the regulation involved in the formation of ventricles and importantly the mechanism surrounding its restriction to optimal number and size.
The nervous system is composed of two main parts, the central nervous system (CNS) and peripheral nervous system (PNS). The PNS connects the CNS to limbs, glands, and organs in our body. In fact, great efforts for recapitulating interaction between the CNS and the PNS have been dedicated. However, it is difficult to mimic the interplay between them due to their high complexity. Current progress in the field of brain organoids have concentrated only on generating CNS organoids. Thus, brain organoids mimicking the combined nervous systems have yet to be developed. Recently, neuromuscular organoids (NMOs) were described from hPSC-derived neuromesodermal progenitors (Faustino Martins et al., 2020). NMOs formed functional neuromuscular junctions with central pattern generator-like circuits. Although spinal cord neurons and skeletal muscles developed in parallel and displayed functional innervation in the organoids, the corticospinal tract from the cortex at the organoid level should be addressed in the future. We are not far away from creating comprehensive brain organoids that contains all of the CNS-specific regions along with the PNS. In addition, we need to refine protocols to obtain mature PNS organoids. If successful, these organoids would provide unparalleled tools to understand the cross talk between CNS and PNS in the developing fetal brain.
One of the major limitations of brain organoids is their variability within and between organoid batches, resulting in limited reproducibility. In fact, organoid variability might lead to data misinterpretation such as masking phenotypic discovery between disease and healthy brain organoids. In particular, even in the same batch, cerebral organoids produced through unguided protocols showed different morphologies, sizes, and cytoarchitectures (Lancaster and Knoblich, 2014; Lancaster et al., 2013). One of the main reasons for these differences may be the use of Matrigel. Although Matrigel supports organoid formation and the growth of neuroepithelium, this undefined proteinaceous mixture can produce a source of variability in experimental outcomes (Hughes et al., 2010). For reducing organoid variability, undefined factors such as Matrigel should be minimized. Recently, to overcome the variability and reproducibility issues in cerebral organoids, elongated EBs were generated using polymer microfilaments. The resulting organoids displayed improved tissue architecture and high reproducibility (Lancaster et al., 2017). More recently, uniform cerebral organoids were reproducibly generated using the modified protocol, including removal of Matrigel before spinning culture and normalizing EB morphogenesis (EB size, optimal treatment of FGF, and well geometry and coatings). Cerebral organoids generated through this protocol exhibited extensive cellular diversity but had similar proportions of cell types across different batches (Sivitilli et al., 2020). Compared with unguided protocols, cortical organoid generated by guided protocols are highly reliable and consistent in terms of cellular composition (Velasco et al., 2019; Yoon et al., 2019). However, heterogeneous cellular responses to patterning signals might contribute to asynchronous differentiation, leading to organoid variability (Miranda et al., 2015). Recently, bioengineering strategy to control aggregate size and stem cell differentiation was applied for improving reproducibility (Zhu et al., 2017). Overall, during organoid generation, uniform size, uniform morphology, and synchronized differentiation of EBs should be considered.
Concluding remarks and future perspectives
Brain organoids have a great potential to bridge the gap between in vivo and in vitro 2D models by mimicking many key features of the human brain. Over the past few years, numerous approaches for generating brain organoids have been established. Typically, cerebral organoids generated by unguided protocols exhibit diverse cellular identities, whereas guided organoids display specific brain regional identities with less heterogeneity (Quadrato et al., 2017; Velasco et al., 2019). Owing to these differences, there is a continuous debate among researchers regarding which approach better serves the research goals. On one hand, cerebral organoids are highly complex and more heterogeneous, compared with brain region-specific organoids. This high heterogeneity might hinder the quantitative analyses, thus the application of cerebral organoids technology to high-throughput screening will be challenging. On the other hand, brain region-specific organoids exhibit relatively homogeneous structures with uniform size, rendering this approach more suited for drug screening and disease modeling platforms. The trade-off between diversity and consistency is a very valid issue in this field. Thus, further defined brain organoid models that satisfy both diversity and consistency are highly desired, but in the end the model of choice is purely dependent on the specific scope of study (Qian et al., 2019).
As with any other evolving technologies, the approaches to analyze brain organoids must also be further refined. Structural, molecular, and functional aspects of brain organoids will require intense investigation. In addition to imaging systems, tissue clearing techniques have helped to obtain better contrasted images with efficient staining in 3D structure (Chung and Deisseroth, 2013). Furthermore, the transcriptional landscape of brain organoids has been revealed at the single-cell level. scRNA-seq allows a better understanding of cellular heterogeneity within brain organoids and provides a valuable resource to identify molecular similarities between brain organoids and fetal brain. In general, analysis of electrophysiological features is a basic and essential requirement for the assessment of neuronal function. Although several approaches such as patch-clamping or calcium imaging have demonstrated that brain organoids are functional, cell type-specific neuronal activity has not been addressed well. Overall, technological improvement and innovation for analyzing brain organoids is still required. Combination of advanced techniques such as Patch-seq, scRNA-seq with patch-clamp recording and morphological analysis, will be one of the options (van den Hurk et al., 2018).
Brain organoids technology offers a new platform for understanding brain development, modeling neurological disease, and drug discovery. To move forward into clinical applications, existing shortcomings of brain organoid technology discussed earlier need to be overcome. Besides, the limited size of brain organoids, cortical arealization, and gyrification should be addressed in further studies. Nevertheless, brain organoids derived from induced pluripotent stem cells (iPSCs) have enormous potentials for precision medicine that identify personalized therapeutic strategies for patients. A combination of brain organoids with advanced techniques such as genome editing will take brain organoid technology to the next level.
Acknowledgments
I.-H. P. was partly supported by NIH (GM111667–01, R01MH118344-01A1, R01MH118554-01A1, R01AA025080–01, R01CA203011-2), CSCRF (14-SCC-YALE-01, 16-RMB-YALE-04), Kavli Foundation, Simons Foundation, and NOMIS Foundation. G.J.S was partly supported by the Norwegian Research Council through its Centers of Excellence funding scheme (project number 262613).
Author contributions
All the authors wrote and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- Amiri A., Coppola G., Scuderi S., Wu F., Roychowdhury T., Liu F., Pochareddy S., Shin Y., Safi A., Song L. Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science. 2018;362:eaat6720. doi: 10.1126/science.aat6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley J.A., Reumann D., Bian S., Levi-Strauss J., Knoblich J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri A., Andrews M.G., Mancia Leon W., Jung D., Shin D., Allen D., Jung D., Schmunk G., Haeussler M., Salma J. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–148. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F., Andersen J., Makinson C.D., Islam S., Wei W., Huber N., Fan H.C., Metzler K.R.C., Panagiotakos G., Thom N. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I., Blakemore C., Rakic P. Development of the human cerebral cortex: boulder Committee revisited. Nat. Rev. Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Cakir B., Xiang Y., Tanaka Y., Kural M.H., Parent M., Kang Y.J., Chapeton K., Patterson B., Yuan Y., He C.S. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods. 2019;16:1169–1175. doi: 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Brauninger M., Lewitus E., Sykes A., Hevers W., Lancaster M. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. U S A. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang X., Peng G. SCSA: a cell type Annotation tool for single-cell RNA-seq data. Front. Genet. 2020;11:490. doi: 10.3389/fgene.2020.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K., Deisseroth K. CLARITY for mapping the nervous system. Nat. Methods. 2013;10:508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- Dekkers J.F., Alieva M., Wellens L.M., Ariese H.C.R., Jamieson P.R., Vonk A.M., Amatngalim G.D., Hu H., Oost K.C., Snippert H.J.G. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 2019;14:1756–1771. doi: 10.1038/s41596-019-0160-8. [DOI] [PubMed] [Google Scholar]
- Durens M., Nestor J., Williams M., Herold K., Niescier R.F., Lunden J.W., Phillips A.W., Lin Y.C., Dykxhoorn D.M., Nestor M.W. High-throughput screening of human induced pluripotent stem cell-derived brain organoids. J. Neurosci. Methods. 2020;335:108627. doi: 10.1016/j.jneumeth.2020.108627. [DOI] [PubMed] [Google Scholar]
- Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M., Wataya T., Nishiyama A., Muguruma K., Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Ekiz H.A., Conley C.J., Stephens W.Z., O'Connell R.M. CIPR: a web-based R/shiny app and R package to annotate cell clusters in single cell RNA sequencing experiments. BMC Bioinformatics. 2020;21:191. doi: 10.1186/s12859-020-3538-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair S.R., Julian D., Hartlaub A.M., Pusuluri S.T., Malik G., Summerfied T.L., Zhao G., Hester A.B., Ackerman W.E.t., Hollingsworth E.W. Electrophysiological maturation of cerebral organoids correlates with dynamic morphological and cellular development. Stem Cell Rep. 2020;15:855–868. doi: 10.1016/j.stemcr.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino Martins J.M., Fischer C., Urzi A., Vidal R., Kunz S., Ruffault P.L., Kabuss L., Hube I., Gazzerro E., Birchmeier C. Self-Organizing 3D human trunk neuromuscular organoids. Cell Stem Cell. 2020;26:172–186 e176. doi: 10.1016/j.stem.2019.12.007. [DOI] [PubMed] [Google Scholar]
- Fuccillo M., Rallu M., McMahon A.P., Fishell G. Temporal requirement for hedgehog signaling in ventral telencephalic patterning. Development. 2004;131:5031–5040. doi: 10.1242/dev.01349. [DOI] [PubMed] [Google Scholar]
- Giandomenico S.L., Mierau S.B., Gibbons G.M., Wenger L.M.D., Masullo L., Sit T., Sutcliffe M., Boulanger J., Tripodi M., Derivery E. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 2019;22:669–679. doi: 10.1038/s41593-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs H.C., Chang-Gonzalez A., Hwang W., Yeh A.T., Lekven A.C. Midbrain-hindbrain boundary morphogenesis: at the intersection of Wnt and fgf signaling. Front. Neuroanat. 2017;11:64. doi: 10.3389/fnana.2017.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C., Konnerth A. Imaging calcium in neurons. Neuron. 2012;73:862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Guo N.N., Liu L.P., Zheng Y.W., Li Y.M. Inducing human induced pluripotent stem cell differentiation through embryoid bodies: a practical and stable approach. World J. Stem Cells. 2020;12:25–34. doi: 10.4252/wjsc.v12.i1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C.S., Postovit L.M., Lajoie G.A. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- Jo J., Xiao Y., Sun A.X., Cukuroglu E., Tran H.D., Goke J., Tan Z.Y., Saw T.Y., Tan C.P., Lokman H. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell. 2016;19:248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoshima T., Sakaguchi H., Nakano T., Soen M., Ando S., Eiraku M., Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. U S A. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.S., Lee J.S., Leem J.W., Huh Y.J., Kim J.Y., Kim H.S., Park I.H., Daley G.Q., Hwang D.Y., Kim D.W. Robust enhancement of neural differentiation from human ES and iPS cells regardless of their innate difference in differentiation propensity. Stem Cell Rev. Rep. 2010;6:270–281. doi: 10.1007/s12015-010-9138-1. [DOI] [PubMed] [Google Scholar]
- Kim H., Xu R., Padmashri R., Dunaevsky A., Liu Y., Dreyfus C.F., Jiang P. Pluripotent stem cell-derived cerebral organoids reveal human oligodendrogenesis with dorsal and ventral origins. Stem Cell Rep. 2019;12:890–905. doi: 10.1016/j.stemcr.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus J., Kanton S., Kyrousi C., Ayo-Martin A.C., Di Giaimo R., Riesenberg S., O'Neill A.C., Camp J.G., Tocco C., Santel M. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat. Med. 2019;25:561–568. doi: 10.1038/s41591-019-0371-0. [DOI] [PubMed] [Google Scholar]
- Kojima T., Hirota Y., Ema M., Takahashi S., Miyoshi I., Okano H., Sawamoto K. Subventricular zone-derived neural progenitor cells migrate along a blood vessel scaffold toward the post-stroke striatum. Stem Cells. 2010;28:545–554. doi: 10.1002/stem.306. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Corsini N.S., Wolfinger S., Gustafson E.H., Phillips A.W., Burkard T.R., Otani T., Livesey F.J., Knoblich J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Muffat J., Omer A., Bosch I., Lancaster M.A., Sur M., Gehrke L., Knoblich J.A., Jaenisch R. Induction of expansion and folding in human cerebral organoids. Cell Stem Cell. 2017;20:385–396 e383. doi: 10.1016/j.stem.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bendito G., Molnar Z. Thalamocortical development: how are we going to get there? Nat. Rev. Neurosci. 2003;4:276–289. doi: 10.1038/nrn1075. [DOI] [PubMed] [Google Scholar]
- Lun M.P., Monuki E.S., Lehtinen M.K. Development and functions of the choroid plexus-cerebrospinal fluid system. Nat. Rev. Neurosci. 2015;16:445–457. doi: 10.1038/nrn3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan M., Nevin Z.S., Shick H.E., Garrison E., Clarkson-Paredes C., Karl M., Clayton B.L.L., Factor D.C., Allan K.C., Barbar L. Induction of myelinating oligodendrocytes in human cortical spheroids. Nat. Methods. 2018;15:700–706. doi: 10.1038/s41592-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour A.A., Goncalves J.T., Bloyd C.W., Li H., Fernandes S., Quang D., Johnston S., Parylak S.L., Jin X., Gage F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018;36:432–441. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J., Coppola G., Zhang P., Abyzov A., Provini L., Tomasini L., Amenduni M., Szekely A., Palejev D., Wilson M. FOXG1-Dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton R.M., Miura Y., Sloan S.A., Li Q., Revah O., Levy R.J., Huguenard J.R., Pasca S.P. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 2019;22:484–491. doi: 10.1038/s41593-018-0316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda C.C., Fernandes T.G., Pascoal J.F., Haupt S., Brustle O., Cabral J.M., Diogo M.M. Spatial and temporal control of cell aggregation efficiently directs human pluripotent stem cells towards neural commitment. Biotechnol. J. 2015;10:1612–1624. doi: 10.1002/biot.201400846. [DOI] [PubMed] [Google Scholar]
- Muguruma K., Nishiyama A., Kawakami H., Hashimoto K., Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015;10:537–550. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- Muraro M.J., Dharmadhikari G., Grun D., Groen N., Dielen T., Jansen E., van Gurp L., Engelse M.A., Carlotti F., de Koning E.J. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016;3:385–394 e383. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill A.C., Kyrousi C., Klaus J., Leventer R.J., Kirk E.P., Fry A., Pilz D.T., Morgan T., Jenkins Z.A., Drukker M. A primate-specific isoform of PLEKHG6 regulates neurogenesis and neuronal migration. Cell Rep. 2018;25:2729–2741 e2726. doi: 10.1016/j.celrep.2018.11.029. [DOI] [PubMed] [Google Scholar]
- Ouhaz Z., Fleming H., Mitchell A.S. Cognitive functions and neurodevelopmental disorders involving the prefrontal cortex and mediodorsal thalamus. Front. Neurosci. 2018;12:33. doi: 10.3389/fnins.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.Y., O'Rourke N.A., Nguyen K.D. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Bonfio C., Chadwick J., Begum F., Skehel M., Lancaster M.A. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science. 2020;369:eaaz5626. doi: 10.1126/science.aaz5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham M.T., Pollock K.M., Rose M.D., Cary W.A., Stewart H.R., Zhou P., Nolta J.A., Waldau B. Generation of human vascularized brain organoids. Neuroreport. 2018;29:588–593. doi: 10.1097/WNR.0000000000001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli D., Magliaro C., Ahluwalia A. Experimental and computational methods for the study of cerebral organoids: a review. Front. Neurosci. 2019;13:162. doi: 10.3389/fnins.2019.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polioudakis D., de la Torre-Ubieta L., Langerman J., Elkins A.G., Shi X., Stein J.L., Vuong C.K., Nichterwitz S., Gevorgian M., Opland C.K. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron. 2019;103:785–801 e788. doi: 10.1016/j.neuron.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen A.A., Bhaduri A., Andrews M.G., Nowakowski T.J., Meyerson O.S., Mostajo-Radji M.A., Di Lullo E., Alvarado B., Bedolli M., Dougherty M.L. Establishing cerebral organoids as models of human-specific brain evolution. Cell. 2019;176:743–756 e717. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin J.F., Tasic B., Hjerling-Leffler J., Trimarchi J.M., Awatramani R. Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci. 2016;19:1131–1141. doi: 10.1038/nn.4366. [DOI] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Song H., Ming G.L. Brain organoids: advances, applications and challenges. Development. 2019;146:dev166074. doi: 10.1242/dev.166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Min Yang S., Berger D.R., Maria N., Scholvin J., Goldman M., Kinney J.P. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner M., Lancaster M.A., Bian S., Choi H., Ku T., Peer A., Chung K., Knoblich J.A. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36:1316–1329. doi: 10.15252/embj.201694700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi H., Kadoshima T., Soen M., Narii N., Ishida Y., Ohgushi M., Takahashi J., Eiraku M., Sasai Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X., Liao J., Lu X., Xue R., Ai N., Fan X. scCATCH: automatic annotation on cell types of clusters from single-cell RNA sequencing data. iScience. 2020;23:100882. doi: 10.1016/j.isci.2020.100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Sun L., Wang M., Liu J., Zhong S., Li R., Li P., Guo L., Fang A., Chen R. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 2020;18:e3000705. doi: 10.1371/journal.pbio.3000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitilli A.A., Gosio J.T., Ghoshal B., Evstratova A., Trcka D., Ghiasi P., Hernandez J.J., Beaulieu J.M., Wrana J.L., Attisano L. Robust production of uniform human cerebral organoids from pluripotent stem cells. Life Sci. Alliance. 2020;3:e202000707. doi: 10.26508/lsa.202000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan S.A., Darmanis S., Huber N., Khan T.A., Birey F., Caneda C., Reimer R., Quake S.R., Barres B.A., Pasca S.P. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron. 2017;95:779–790 e776. doi: 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A.M.M., Meyer K.A., Santpere G., Gulden F.O., Sestan N. Evolution of the human nervous system function, structure, and development. Cell. 2017;170:226–247. doi: 10.1016/j.cell.2017.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambalo M., Lodato S. Brain organoids: human 3D models to investigate neuronal circuits assembly, function and dysfunction. Brain Res. 2020;1746:147028. doi: 10.1016/j.brainres.2020.147028. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Cakir B., Xiang Y., Sullivan G.J., Park I.H. Synthetic analyses of single-cell transcriptomes from multiple brain organoids and fetal brain. Cell Rep. 2020;30:1682–1689 e1683. doi: 10.1016/j.celrep.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Trujillo C.A., Gao R., Negraes P.D., Gu J., Buchanan J., Preissl S., Wang A., Wu W., Haddad G.G., Chaim I.A. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell. 2019;25:558–569 e557. doi: 10.1016/j.stem.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk M., Erwin J.A., Yeo G.W., Gage F.H., Bardy C. Patch-seq protocol to analyze the electrophysiology, morphology and transcriptome of whole single neurons derived from human pluripotent stem cells. Front. Mol. Neurosci. 2018;11:261. doi: 10.3389/fnmol.2018.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Buth J.E., Vishlaghi N., de la Torre-Ubieta L., Taxidis J., Khakh B.S., Coppola G., Pearson C.A., Yamauchi K., Gong D. Self-organized cerebral organoids with human-specific features predict effective drugs to combat zika virus infection. Cell Rep. 2017;21:517–532. doi: 10.1016/j.celrep.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Tanaka Y., Cakir B., Patterson B., Kim K.Y., Sun P., Kang Y.J., Zhong M., Liu X., Patra P. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell. 2019;24:487–497 e487. doi: 10.1016/j.stem.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Tanaka Y., Patterson B., Kang Y.J., Govindaiah G., Roselaar N., Cakir B., Kim K.Y., Lombroso A.P., Hwang S.M. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21:383–398 e387. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Yoshiaki T., Patterson B., Cakir B., Kim K.Y., Cho Y.S., Park I.H. Generation and fusion of human cortical and medial ganglionic eminence brain organoids. Curr. Protoc. Stem Cell Biol. 2018;47:e61. doi: 10.1002/cpsc.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaziji H., Barry T. Diagnostic Immunohistochemistry: what can go wrong? Adv. Anat. Pathol. 2006;13:238–246. doi: 10.1097/01.pap.0000213041.39070.2f. [DOI] [PubMed] [Google Scholar]
- Yoon S.J., Elahi L.S., Pasca A.M., Marton R.M., Gordon A., Revah O., Miura Y., Walczak E.M., Holdgate G.M., Fan H.C. Reliability of human cortical organoid generation. Nat. Methods. 2019;16:75–78. doi: 10.1038/s41592-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A., Munoz-Manchado A.B., Codeluppi S., Lonnerberg P., La Manno G., Jureus A., Marques S., Munguba H., He L., Betsholtz C. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zhao C., Deng W., Gage F.H. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Wang L., Yu H., Yin F., Wang Y., Liu H., Jiang L., Qin J. In situ generation of human brain organoids on a micropillar array. Lab Chip. 2017;17:2941–2950. doi: 10.1039/c7lc00682a. [DOI] [PubMed] [Google Scholar]