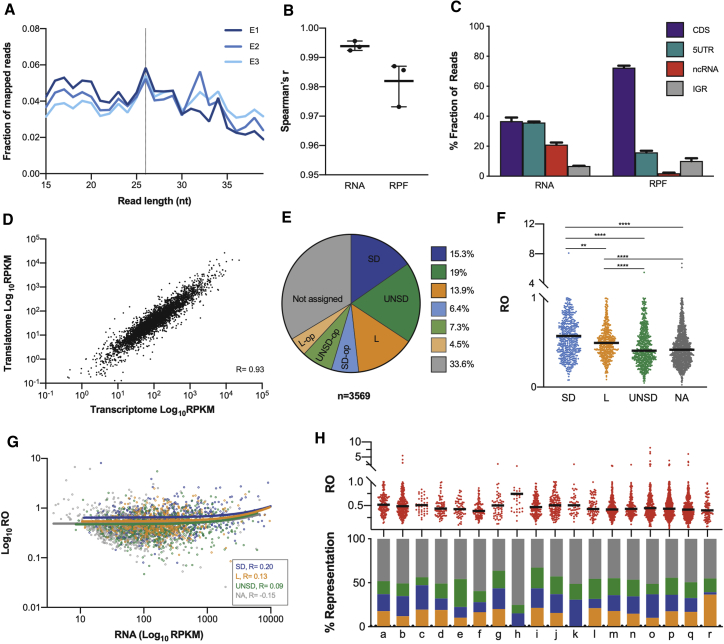
Figure 1.
The translational landscape of M. tuberculosis during exponential growth
(A) Fraction of reads from 15 to 40 nucleotides in M. tuberculosis Ribo-seq libraries during exponential growth (three biological replicates).
(B) Pairwise correlation coefficients calculated between each pair of replicates for total RNA and RPFs based on number of counts per gene. Line and bars represent the mean with range across the three replicates.
(C) Fraction of RNA and RPF reads aligning to various genomic features, coding sequences (CDSs), 5′ UTRs, non-coding RNAs (ncRNA), and intergenic regions (IGRs). Values are represented as percentages. Error bars indicate standard deviation across the three replicates.
(D) Correlation between transcriptome and translatome assessed from plotting average RPKMs across the three replicates for mRNA and RPFs.
(E) Distribution of gene categories among the 3,569 genes for which reliable rates of translation were quantified. Shine-Dalgarno (SD, blue), leaderless (L, orange), 5′ UTR not including a Shine-Dalgarno (UNSD, green), and not assigned (gray). The suffix “op” refers to genes assigned to operons.
(F) Plot of ribosome occupancy (RO) values distribution of Shine-Dalgarno (SD, blue), leaderless (L, orange), UNSD (green), and not assigned (NA) genes during exponential growth. Bars denote median RO values for each group. Asterisk denotes significant differences between gene categories (Kruskal-Wallis test, p < 0.0018).
(G) Plots of mRNA versus RO with Spearman coefficients and linear regression included for each gene category.
(H) Top: RO values distribution among functional categories (see key below). Bars indicate median RO values. Bottom: proportion of Shine-Dalgarno (blue), leaderless (orange), UNSD (green), or not assigned (gray) genes representing each functional category. Key to functional categories: a, degradation; b, energy metabolism; c, central intermediary metabolism; d, amino acid biosynthesis; e, purines, pyrimidines, nucleosides, and nucleotides; f, biosynthesis of cofactors, prosthetic groups, and carriers; g, lipid biosynthesis; h, polyketide and non-ribosomal peptide synthesis; i, broad regulatory functions; j, synthesis and modification of macromolecules; k, ribosomal proteins; l, degradation of macromolecules; m, cell envelope; n, cell processes; o, other; p, conserved hypotheticals; q, unknowns; r, toxin-antitoxin systems.
See also Figures S1 and S2 and Tables S1 and S2.