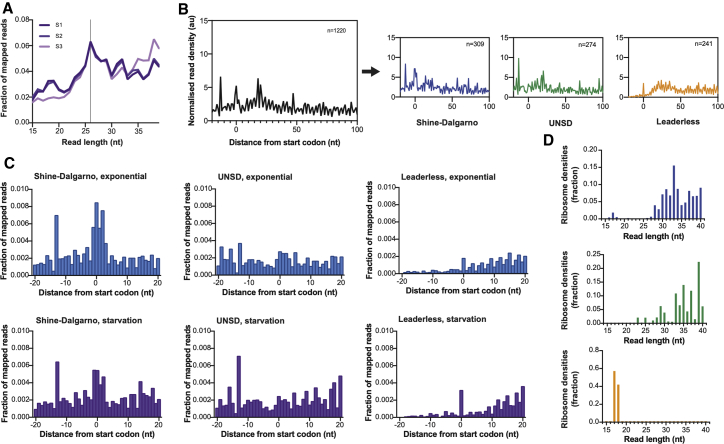
Figure 3.
Mechanisms of translation in starvation
(A) Fraction of reads from 15 to 40 nucleotides in M. tuberculosis Ribo-seq libraries during nutrient starvation (three biological replicates).
(B) Average ribosome density on genes aligned at the start codon during nutrient starvation. Metagene maps for all genes are shown in black, Shine-Dalgarno genes in blue, UNSD genes in green, and leaderless genes in orange. For all maps, the number of genes included in the analysis is indicated.
(C) Fraction of mapped reads around the start codon for Shine-Dalgarno, UNSD, and leaderless genes during exponential growth (blue) and nutrient starvation (purple).
(D) Distribution of RPF read lengths of initiating ribosomes for each gene category: Shine-Dalgarno, blue; UNSD, green; leaderless, orange.
See also Figures S3 and S4.