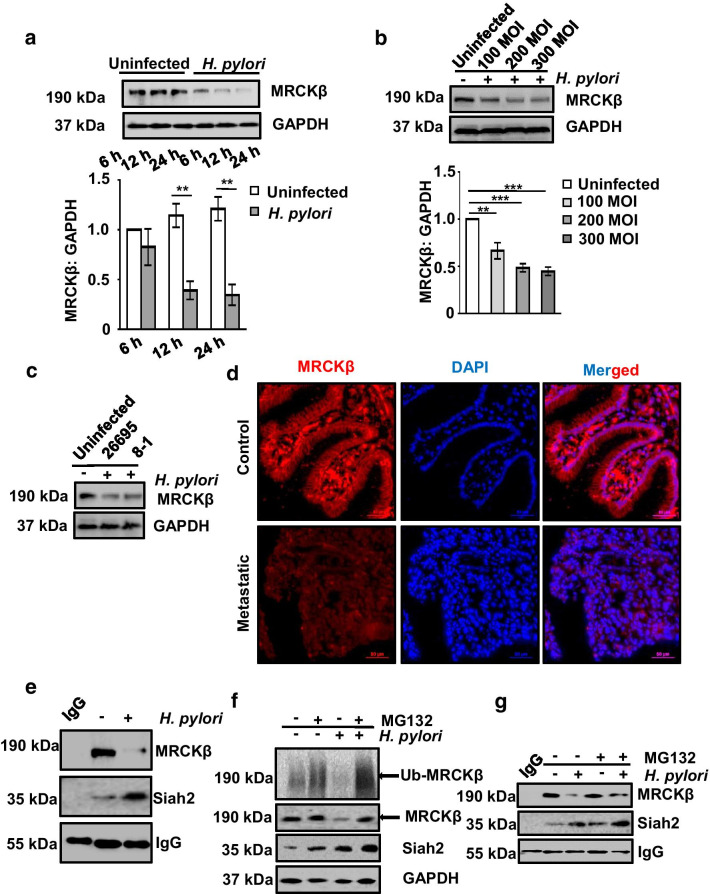
Fig. 2.
MRCKβ is decreased in H. pylori-mediated GC and interacts with Siah2. a Western blot of whole cell extracts from uninfected and H. pylori-infected MKN45 cells (at 6, 12, and 24 h of infection with 200 MOI of H. pylori 26695) showing protein level of MRCKβ. GAPDH is a loading control. Bar graph clearly indicates a time-dependent decrease of MRCKβ after H. pylori infection. Two-way ANOVA followed by Tukey’s post hoc analysis is used to determine statistical significance. b Western blotting of total cell lysate from uninfected and H. pylori-infected (at MOI 100, 200 and 300) MKN45 cells showing MRCKβ protein. GAPDH is used as a loading control. Graph represents decrease of MRCKβ protein as compared to uninfected control. One-way ANOVA is used to determine statistical significance. c A representative western blot of infected (200 MOI for 12 h of H. pylori 26695 or 8–1 strain) or uninfected MKN45 cells showing MRCKβ protein level (n = 3). GAPDH is used as a loading control. d A representative (n = 9) immunofluorescence micrograph of human metastatic GC biopsy tissue samples showing the status of MRCKβ. Tissues are sectioned at 5 μm thickness. Images are captured using 40X objective and scale bars = 50 μm. e Western blot of whole cell lysate from uninfected and infected (H. pylori at 200 MOI for 12 h) MKN45 cells subjected to co-immunoprecipitation using Siah2 antibody reveals that Siah2 interacts with MRCKβ. IgG band is used to indicate equal loading. f A representative western analysis indicating ubiquitination status of MRCKβ from uninfected or infected (H. pylori at 200 MOI for 12 h) and 50 µM MG132-treated MKN45 cells. Reprobed bands of MRCKβ, Siah2 and GAPDH. g Western blotting of total cell lysates from uninfected or infected (H. pylori at 200 MOI for 12 h) and 50 µM MG132-treated MKN45 cells subjected to co-immunoprecipitation using Siah2 antibody showing MRCKβ and Siah2 protein status. IgG band is used as the indicator of equal loading. All data are mean ± sem (n = 3). **P < 0.01, ***P < 0.001