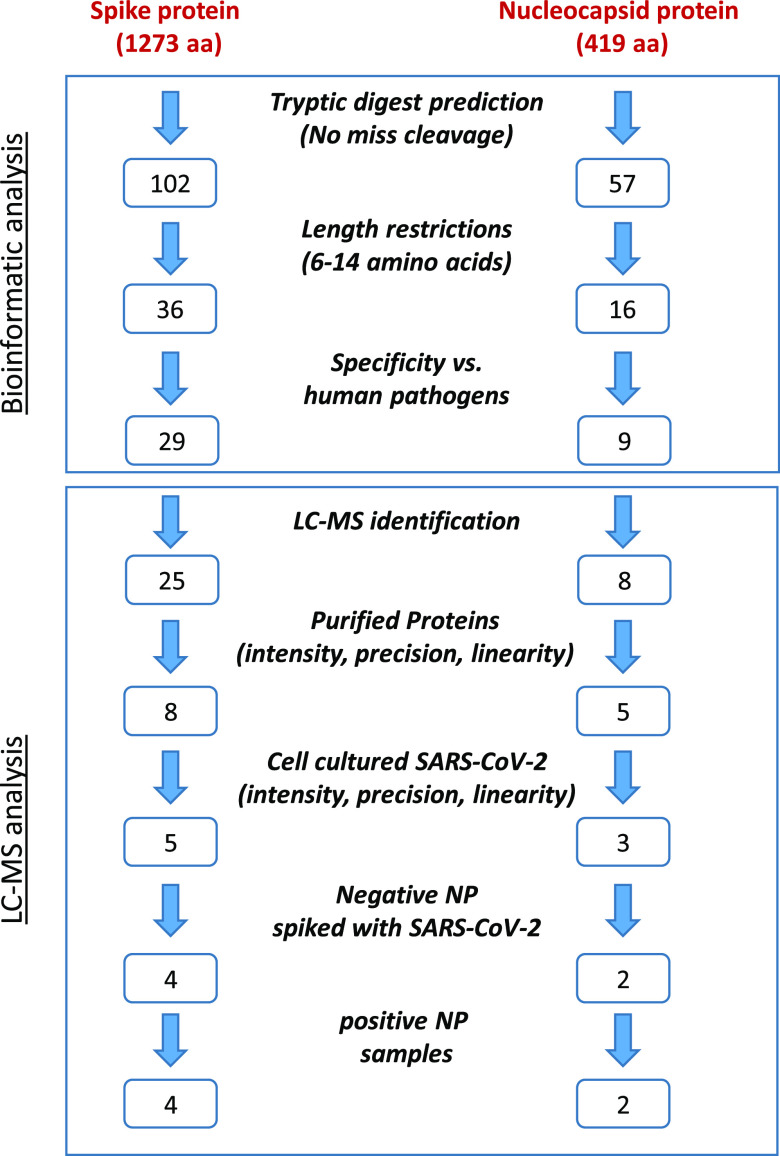
Figure 1.
Methodology flowchart for SARS-CoV-2 marker selection. The first part was performed computationally, including in silico tryptic digest followed by length restriction (6–14 aa) and specificity consideration (removal of peptides that appear in human pathogen databases). The second part is LC-MS analysis of tryptic peptides derived from purified proteins, cell-cultured SARS-CoV-2, negative NP sample spiked with SARS-CoV-2, and positive NP samples. Evaluating parameters as sensitivity, reproducibility, and linearity helped reduce the number of potential markers.