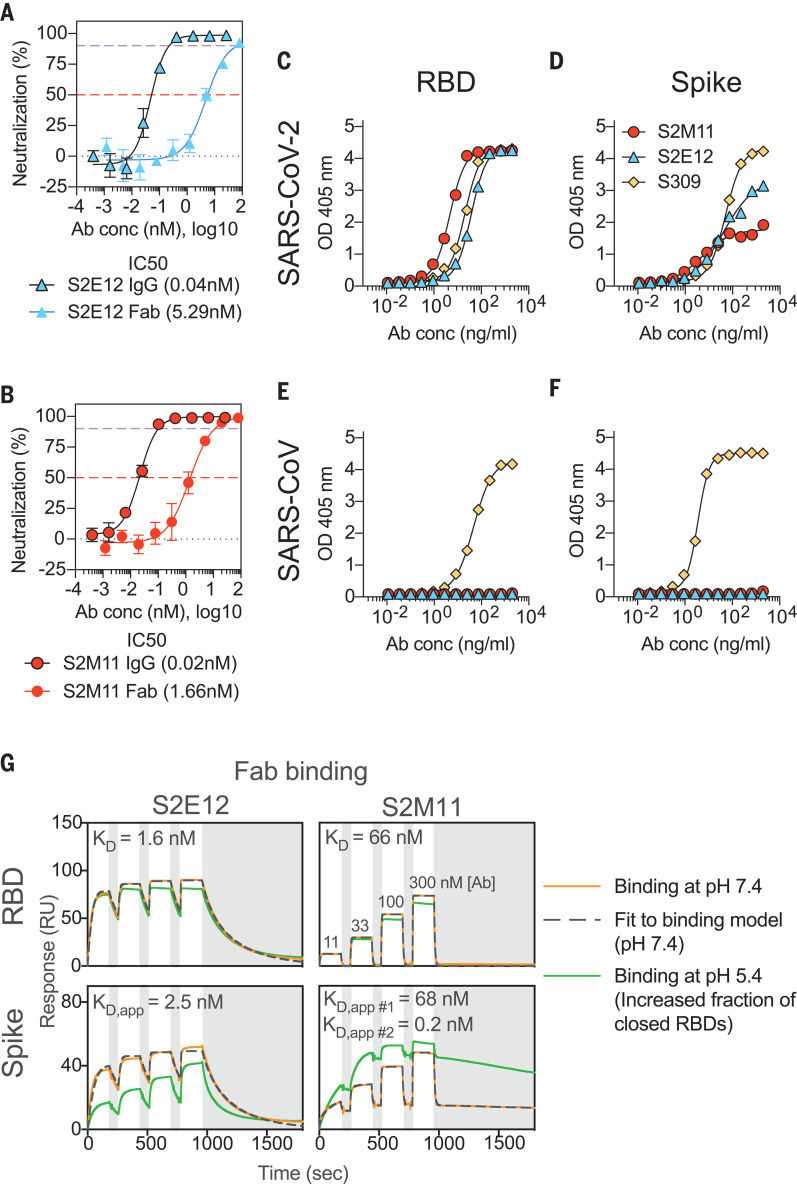
Fig. 1. S2E12 and S2M11 neutralize SARS-CoV-2 ultrapotently by targeting the RBD.
(A and B) Neutralization of authentic SARS-CoV-2 (SARS-CoV-2-Nluc) by S2E12 (A) and S2M11 (B) IgG or Fab. Symbols are means ± SD of triplicates. Dashed lines indicate IC50 and IC90 values. Average IC50 values are indicated in parentheses below the graphs (determined from two independent experiments). (C to F) ELISA binding of S2M11 (red), S2E12 (blue), or S309 (yellow) mAbs to immobilized SARS-CoV-2 RBD (C), SARS-CoV-2 S (D), SARS-CoV RBD (E), or SARS-CoV S (F). Symbols show means of duplicates. (G) SPR analysis of S2E12 and S2M11 Fab binding to the SARS-CoV-2 RBD or S ectodomain trimer. Experiments were carried out at pH 7.4 (orange) and pH 5.4 (green) and were repeated twice with similar results (one experiment is shown). The apparent equilibrium dissociation constants (KD, app) at pH 7.4 are indicated. White and gray stripes indicate association and dissociation phases, respectively. S2M11 binding to S was fit to two parallel kinetic phases and the resulting KD, app #1 and KD, app #2 were interpreted as apparent affinities for open RBDs (tertiary epitope) and closed RBDs (quaternary epitope), respectively. This is supported by the similar binding kinetics and affinity of the faster off-rate phase (KD, app #1) with that observed for S2M11 binding to the isolated RBD (compare with table S1 for full fit results). Ab conc, mAb concentration.