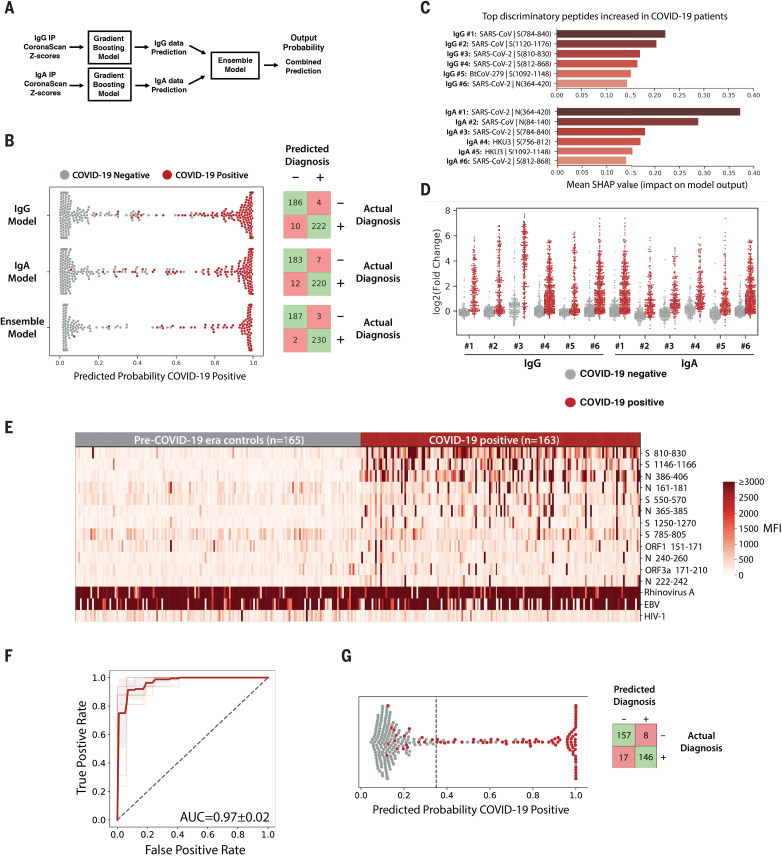
Fig. 4. Machine learning models trained on VirScan data discriminate COVID-19–positive and –negative individuals with very high sensitivity and specificity.
(A) Gradient-boosting machine learning models were trained on IgG and IgA VirScan data from 232 COVID-19 patients and 190 pre–COVID-19 era controls. Separate models were created for the IgG and IgA data, and then a third model (Ensemble) was trained to combine the outputs of the first two. (B) The plot shows the predicted probability that each sample is positive for COVID-19. True COVID-19–positive samples are shown as red dots; true COVID-19–negative samples are shown as gray dots. The corresponding confusion matrix for each model is shown on the right. (C and D) SHAP analysis to identify the most discriminatory peptides informing the models in (B). The chart in (C) summarizes the relative importance of the most discriminatory peptides increased among COVID-19 patients identified by the IgG and IgA gradient-boosting models. The enrichment [log2(fold change) of the normalized read counts in the sample IP versus in no-serum control reactions] of each of these peptides across all samples is shown in (D). (E) Luminex assay using highly discriminatory SARS-CoV-2 peptides identifies IgG antibody responses in COVID-19 patients but rarely in pre–COVID-19 era controls. Each column represents a COVID-19 patient (n = 163) or pre–COVID-19 era control (n = 165); each row is a SARS-CoV-2–specific peptide. Peptides containing public epitopes from rhinovirus A, EBV, and HIV-1 served as positive and negative controls. The color scale indicates the median fluorescence intensity (MFI) signals after background subtraction. (F) Receiver operating characteristic (ROC) curve for the Luminex assay predicting SARS-CoV-2 infection history, evaluated by 10× cross-validation. The light red lines indicate the ROC curve for each test set, the dark line indicates the average, and the gray region represents ±1 SD. The average area under the curve (AUC) is shown. (G) (Left) Predicted probability that each sample is positive for COVID-19, using the Luminex model, as in (B). The dashed line indicates the model threshold. (Right) Confusion matrix for the Luminex model.