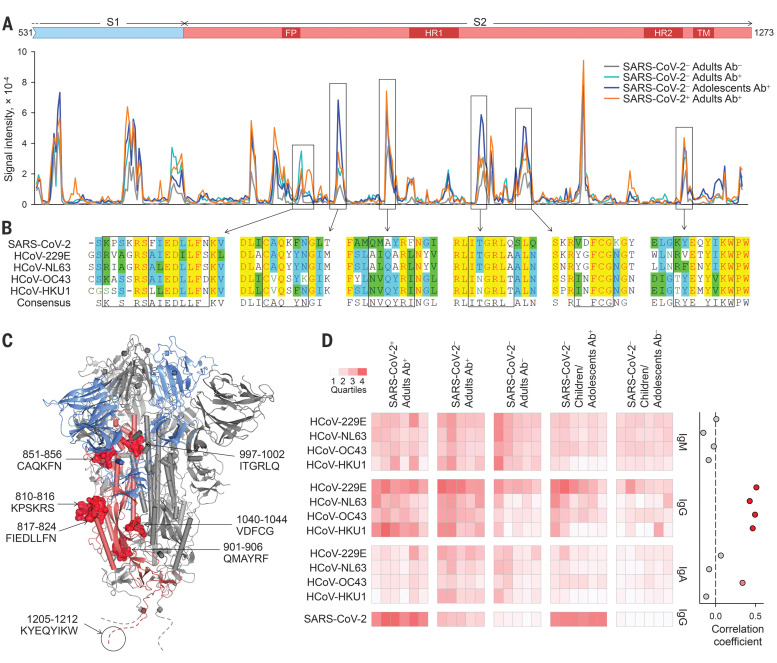
Fig. 4. Mapping of cross-reactive epitopes in SARS-CoV-2 S.
(A) Signal intensity for each overlapping peptide along the length of SARS-CoV-2 S covered in the peptide arrays using pooled sera with (Ab+) or without (Ab−) flow cytometry–detectable SARS-CoV-2 S–reactive antibodies. Differentially recognized peaks are boxed. (B) Alignment of the amino acid sequences of SARS-CoV-2 and HCoV S glycoproteins. Boxes indicate predicted core epitopes. (C) Mapping of predicted epitopes targeted on the trimeric SARS-CoV-2 spike. The S1 (blue) and S2 (pink) subunits of one monomer are colored. Epitopes are shown for one monomer; the circled dashed line represent the membrane proximal region not present in the structure. (D) Left: Reactivity with the S glycoproteins of each HCoV of the indicated sera with (Ab+) or without (Ab−) flow cytometry–detectable SARS-CoV-2 S–reactive antibodies as determined by flow cytometry. Each column is an individual sample. Rows depict the staining for each antibody class. Right: Correlation coefficients between percentages of IgG staining for SARS-CoV-2 S and IgG, IgM, and IgA staining for each HCoV S glycoprotein.