The protease kallikrein 13 cleaves the spike protein of the human coronavirus HKU1 to facilitate viral entry.
Priming viral entry
Unlike SARS-CoV-2, the human coronavirus HKU1 normally causes relatively mild respiratory tract infections; however, it shares with SARS-CoV-2 the mechanism of using its surface spike (S) protein to enter target cells. Because the host receptor for HCoV-HKU1 is unknown, efforts to study the virus in cell culture systems have proved difficult. Milewska et al. found that knockout of the protease kallikrein 13 (KLK13) in human airway epithelial cells blocked their infection by HCoV-HKU1, that overexpression of KLK13 in nonpermissive cells enabled their infection by the virus, and that KLK13 cleaved the viral S protein. Together, these findings suggest that KLK13 is a priming enzyme for viral entry and may help to establish cell lines that can facilitate further investigation of the mechanism of viral pathogenesis.
Abstract
Human coronavirus HKU1 (HCoV-HKU1) is associated with respiratory disease and is prevalent worldwide, but an in vitro model for viral replication is lacking. An interaction between the coronaviral spike (S) protein and its receptor is the primary determinant of tissue and host specificity; however, viral entry is a complex process requiring the concerted action of multiple cellular elements. Here, we found that the protease kallikrein 13 (KLK13) was required for the infection of human respiratory epithelial cells and was sufficient to mediate the entry of HCoV-HKU1 into nonpermissive RD cells. We also demonstrated the cleavage of the HCoV-HKU1 S protein by KLK13 in the S1/S2 region, suggesting that KLK13 is the priming enzyme for this virus. Together, these data suggest that protease distribution and specificity determine the tissue and cell specificity of the virus and may also regulate interspecies transmission.
INTRODUCTION
Coronaviruses are the largest group within the order Nidovirales. Mainly, they cause respiratory and enteric diseases in humans and animals, but some can cause more severe conditions, such as hepatitis, peritonitis, or neurological disease. Seven coronaviruses infect humans, four of which [human coronavirus (HCoV)–229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1] cause relatively mild upper and lower respiratory tract disease, and two [severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV)] are associated with severe, life-threatening respiratory infections and multiorgan failure (1–6). Furthermore, in December 2019, a previously uncharacterized coronavirus emerged in Wuhan City, China, causing pneumonia. To date, this virus (named SARS-CoV-2) has spread worldwide, causing a global pandemic with millions of confirmed cases and almost hundreds of thousands of fatalities reported (www.cdc.gov/coronavirus/novel-coronavirus-2019.html; https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html).
Coronaviral infection is initiated by an interaction between the trimeric spike (S) protein of the virus and its receptor, which is expressed on the surface of the susceptible cell. Several adhesion and entry receptors have been described for coronaviruses. For example, HCoV-229E (which is similar to many other alphacoronaviruses) uses aminopeptidase N as the primary entry port (7). Unexpectedly, another alphacoronavirus, HCoV-NL63, shares receptor specificity with the evolutionarily distant SARS-CoV, both hijack angiotensin-converting enzyme 2 (ACE2) (8–11). HCoV-NL63 also uses heparan sulfate as a primary attachment site (12–14). A very different receptor is recognized by MERS-CoV, which binds to dipeptidyl-peptidase 4 (9, 15, 16). Another betacoronavirus, HCoV-OC43, binds to N-acetyl-9-O-acetylneuraminic acid (17, 18). HCoV-HKU1 remains the great unknown because its cellular receptor has not yet been identified, and all efforts to culture the virus in vitro have failed.
HCoV-HKU1 was identified in Hong Kong in 2004. The virus was present in a sample obtained from an elderly patient with severe pneumonia (19). Epidemiological studies showed a high prevalence of the pathogen in humans. This is because most children seroconvert before the age of 6 years (20, 21). Although it is not possible to culture HCoV-HKU1 in vitro, we and others reported that ex vivo fully differentiated human airway epithelium (HAE) cultures and human alveolar type II cells support the infection (22–25). A study by Huang et al. (26) demonstrated that HCoV-HKU1 binds to target cells through O-acetylated sialic acids on the cell surface; however, this interaction is not sufficient for infection. This study also showed that the hemagglutinin-esterase (HE) protein of HCoV-HKU1 exhibits sialate-O-acetylesterase activity and may act as a receptor-destroying enzyme, thereby facilitating the release of viral progeny (26). Bakkers et al. (27) proposed that, to adapt to the sialoglycome of the human respiratory tract over the evolutionary time scale, HCoV-HKU1 lost the ability to bind to attachment receptors through the HE protein. Hulswit et al. (28) mapped the virus-binding site to O-acetylated sialic acids, demonstrating that the S1 domain A is responsible for binding to the attachment receptor.
The S protein is the leading player during coronavirus entry, and its characteristics determine the host range. Coronaviral S proteins are class I fusion proteins, consisting of a large N-terminal ectodomain, a hydrophobic transmembrane region, and a small C-terminal endodomain. The ectodomain is highly glycosylated and is composed of S1 and S2 domains. The globular S1 domain is highly variable and carries the receptor-binding site, whereas the more conserved, rod-like S2 domain undergoes structural rearrangement during entry, which brings the cellular and viral membranes into proximity with each other. Such a structural switch may be triggered by different stimuli, including receptor binding, proteolytic cleavage of the S protein, or a reduction in pH. Because different species require different stimuli, coronaviruses enter cells at different subcellular sites. Some coronaviruses fuse at the plasma membrane, whereas others are believed to enter the cell through receptor-mediated endocytosis, which is followed by fusion deep within the endosomal compartments (29–35). Furthermore, reports showed that the entry portal may vary depending on tissue or cell characteristics. These differences may affect the host range, pathogenicity, and cell or tissue specificity (1).
Host proteases prime coronaviral S proteins. For example, trypsin-mediated cleavage of the S protein of porcine epidemic diarrhea virus is required for entry in the small intestine (36), whereas a number of coronaviruses from different genera [including HCoV-OC43, HCoV-HKU1, murine hepatitis virus (MHV), MERS-CoV, and infectious bronchitis virus] have a furin cleavage site (37–41). Kam et al. (42) showed that the SARS-CoV S protein can be cleaved by plasmin; however, there is almost no biological evidence for its role in vivo. Cathepsins may also act as S protein–activating enzymes. Cathepsin L processes the S proteins of SARS-CoV, MERS-CoV, HCoV-229E, and MHV-2 (43–46). However, reports showed that, in vivo, respiratory coronaviruses may be activated by the transmembrane protease serine 2 (TMPRSS2), which enables endocytosis-independent internalization, thereby reshaping the entry process (45, 47–50). Whereas the S proteins of laboratory strains of viruses require priming by cathepsins, the S proteins of clinical isolates (for example, HCoV-229E and HCoV-OC43) undergo TMPRSS2-mediated cleavage at the cell surface, which enables them to fuse with the cell membrane on the cell surface (51). A study by Shirato et al. (51) demonstrated that coronaviruses may lose their ability to infect naturally permissive HAE cultures during cell culture adaptation because the gene encoding the S protein evolves and adjusts to the proteolytic landscape of the immortalized cells.
Here, we identified a protease belonging to the tissue kallikrein (KLK) family as a previously uncharacterized factor essential for HCoV-HKU1 entry to the target cell. The KLK family consists of 15 closely related serine proteases with trypsin- or chymotrypsin-like specificity. The expression of these enzymes is tightly regulated, and each tissue has its own unique KLK expression profile. These enzymes play roles in a diverse range of processes during embryonic development to adulthood (52–56), and some have been linked to human cancers (57–60). The function of some KLKs remains to be elucidated, but the current evidence suggests that protease distribution may be an important factor that not only predetermines the cell and tissue specificity of the virus but also regulates interspecies transfers. Furthermore, the data presented herein bring us a step closer to developing an in vitro cell culture model and possibly identifying the cellular receptor for this virus.
RESULTS
Several KLKs are increased in abundance after infection of HAE cultures with HCoV-HKU1
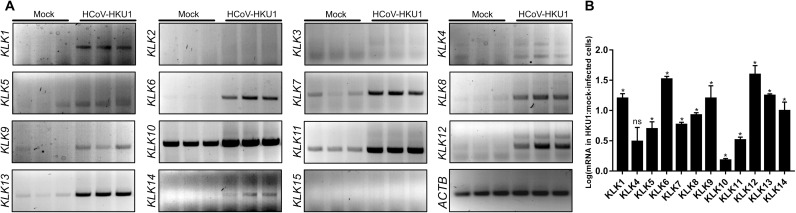
First, we asked whether HCoV-HKU1 infection modulated the expression of human KLKs. HAE cultures were infected with HCoV-HKU1 or mock-inoculated. At 120 hours post-inoculation (p.i.), cells were collected, and the relative abundance of mRNAs encoding KLKs was analyzed. We detected the expression of KLK7, KLK8, KLK10, KLK11, and KLK13 in uninfected, fully differentiated cell cultures (Fig. 1). However, the pattern in HCoV-HKU1–infected cells was different: We detected an increase in the amounts of KLK7, KLK8, KLK10, KLK11, and KLK13 mRNAs. Furthermore, KLK1, KLK5, KLK6, KLK9, KLK12, and KLK14 were expressed in the infected cells, whereas KLK2, KLK3, and KLK15 were not expressed (Fig. 1).
Fig. 1. HCoV-HKU1 infection of HAE cultures induces the expression of several KLKs.
(A and B) HAE cultures were left uninfected (mock) or were infected with HCoV-HKU1 (106 RNA copies/ml) for 2 hours at 32°C and cultured for 5 days. Cellular RNA was then isolated, treated with DNase, and subjected to reverse transcription, and the mRNAs for the indicated KLKs were amplified using specific primers. The analysis was performed twice using cells obtained from different donors, each time in triplicate. (A) The indicated amplified PCR products were resolved and detected in 1.5% (w/v) agarose gel in 1× TAE buffer. (B) The relative abundance of the indicated KLK mRNAs normalized to that of ACTB was assessed semiquantitively by densitometric analysis. Data are presented as a log change of signal specific for the indicated KLK mRNA in HCoV-HKU1–infected cells compared to that in the mock-infected cells. The experiments were performed twice with cells from different donors, each time with two biological replicates. For comparisons by Student’s t test, *P < 0.05; ns, not significant.
KLK13 is essential for infection by HCoV-HKU1
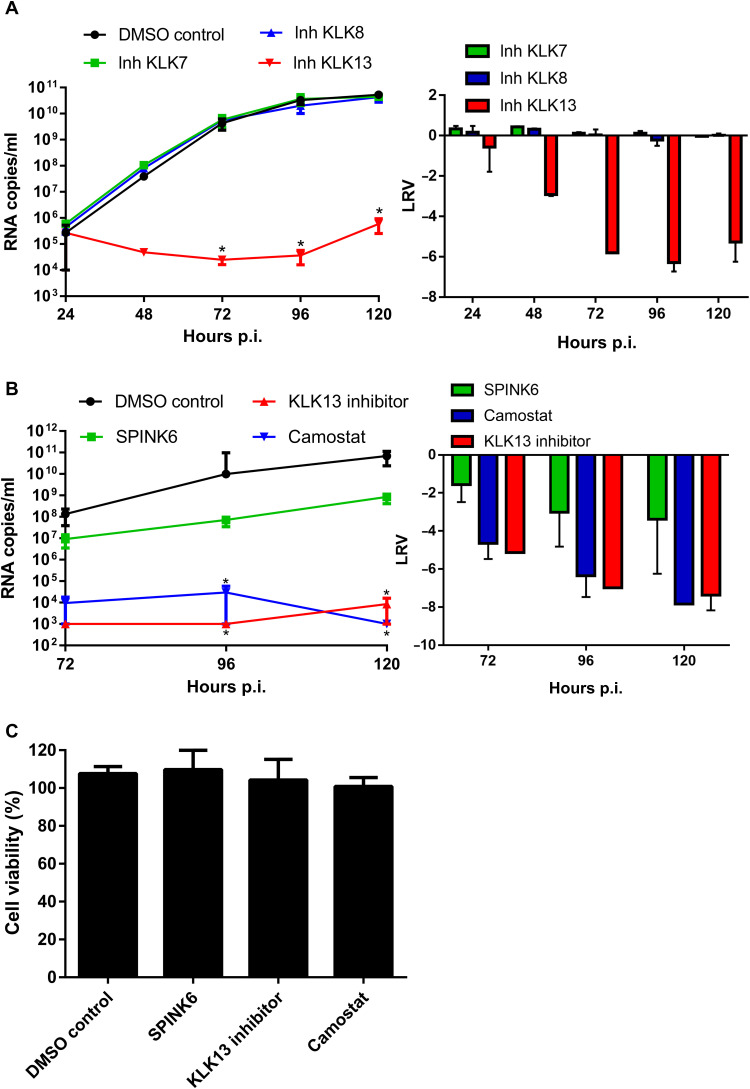
S protein priming is a prerequisite for coronavirus entry; therefore, we tested whether KLKs took part in this process by culturing cells in the presence or absence of KLK inhibitors (table S1) (61). HAE cultures were preincubated with each inhibitor (10 μM) or with vehicle control dimethyl sulfoxide (DMSO) and mock-inoculated or inoculated with the virus (106 RNA copies/ml) in the presence of the inhibitor. Apical washes were collected each day for the analysis of virus replication. Subsequently, viral RNA was isolated and reverse-transcribed, and the HCoV-HKU1 yield was determined by quantitative real-time polymerase chain reaction (qPCR) analysis. The results showed that HCoV-HKU1 replication was inhibited in the presence of a KLK13 inhibitor, which was not the case for cells treated with DMSO or with inhibitors specific for KLK7 or KLK8 (Fig. 2A).
Fig. 2. HCoV-HKU1 infection is dependent on KLK13 activity.
(A and B) HAE cultures were inoculated with HCoV-HKU1 (106 RNA copies/ml) for 2 hours at 32°C in the presence of the indicated KLK inhibitors (each at 10 μM, table S1) or DMSO (A) or else in the presence of SPINK6 (10 μg/ml), 10 μM KLK13 inhibitor, 100 μM camostat, or DMSO. Statistical significance was assessed with the Kruskal-Wallis test. *P < 0.05. (B) To analyze viral replication kinetics, each day post-infection (p.i.), 100 μl of PBS containing a given inhibitor was applied to the apical surface of the HAE cultures and collected after 10 min of incubation at 32°C. Replication of HCoV-HKU1 was evaluated by RT-qPCR analysis, and the data are presented as RNA copy numbers/ml (left) and as the log removal value (LRV) compared to the untreated sample (right). The assays were performed twice, each time in triplicate (N = 3), and average values with standard errors are presented. (C) Assessment of the cytotoxicity of inhibitors in the HAE cultures. Cell viability was assessed with the XTT assay on mock-treated cells at 120 hours p.i. Data on the y axis represent the percentage values obtained for the untreated reference samples. The assays were performed in triplicate (N = 3), and average values with standard errors are presented.
Next, we analyzed HCoV-HKU1 replication in the presence of a family-specific KLK inhibitor SPINK6 at a concentration of 10 μg/ml (62, 63) or 100 μM camostat (a broad inhibitor of serine proteases, also a potent inhibitor of KLK13) (34, 64). We noted inhibition of HCoV-HKU1 replication in the presence of both inhibitors. The effect of SPINK6 was milder than those camostat or the KLK13-specific inhibitor (Fig. 2B), but the 99.9% reduction in virus yield was significant. The pH-dependent stability of SPINK6 and the reversible character of the SPINK6-KLK13 complex might have been a limiting factor. To ensure that the observed inhibitory effects of the inhibitors on viral replication were not due to toxicity, we tested cell viability and found no significant effects (Fig. 2C).
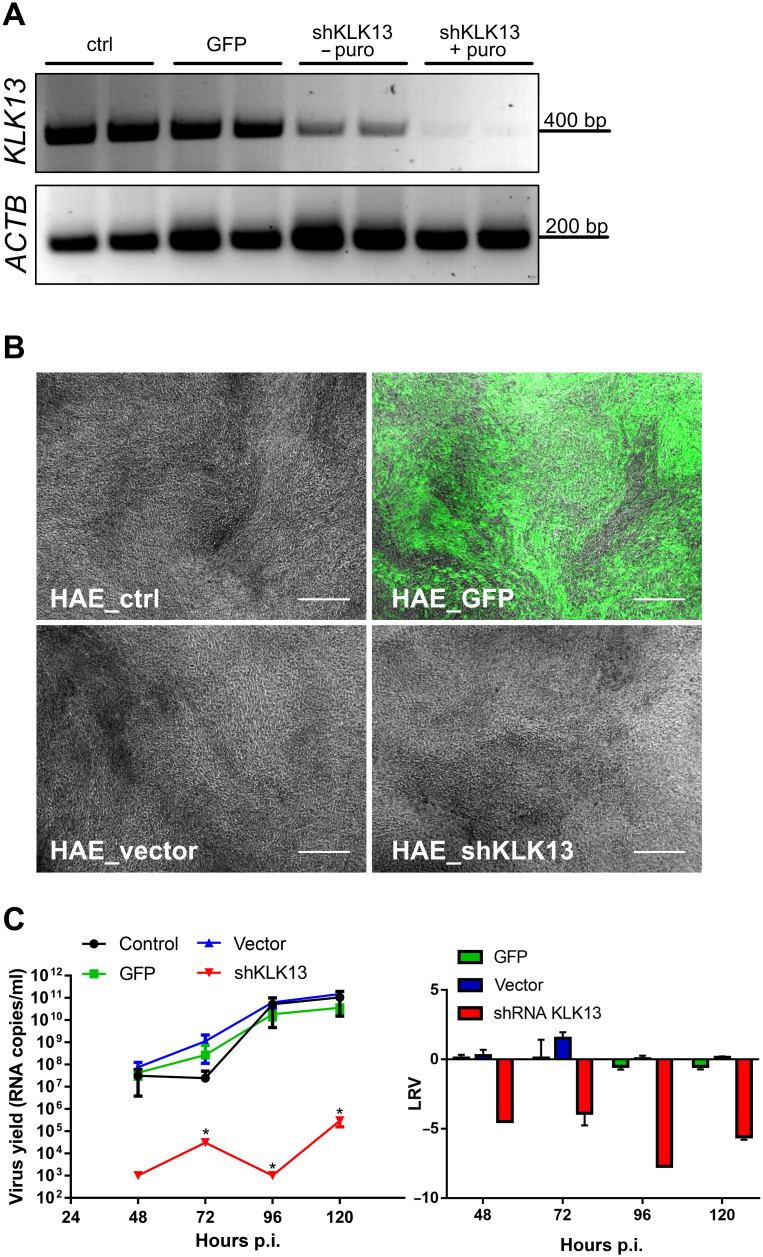
The experiments conducted thus far suggested that KLK13 was required for viral infection. However, one may question the specificity of the KLK13 protease inhibitors. To ensure that KLK13 was indeed the priming enzyme during HCoV-HKU1 infection, we developed HAE cultures with primary cells transduced with lentiviral vectors encoding short hairpin RNAs (shRNAs) targeting KLK13 mRNA. We then confirmed that the expression of the protease was reduced (Fig. 3A, HAE_shKLK13). Nonmodified HAE cultures (HAE_ctrl), cultures transduced with a lentiviral vector to express the green fluorescent protein (HAE_GFP), and HAE cultures transduced with an empty lentiviral vector (HAE_vector) were used as controls. After cellular transduction and differentiation, KLK13 mRNA in HAE_shKLK13 cells was almost undetectable, in contrast to that in the control cultures (Fig. 3A). Furthermore, HAE_shKLK13 cells continued to differentiate and formed pseudostratified cultures (Fig. 3B). Next, we infected HAE_ctrl, HAE_GFP, HAE_vector, and HAE_shKLK13 cells with HCoV-HKU1 (106 RNA copies/ml) and incubated them with the viral stock solution for 2 hours at 32°C. The cultures were maintained at 32°C for 5 days at an air-liquid interface (ALI). Apical washes were collected, and virus yield was determined by reverse transcription (RT)–qPCR analysis. We found that, in contrast to that in control cultures, replication of the virus in the HAE_shKLK13 cells was abolished (Fig. 3C). Together, these data indicate that silencing the KLK13 gene in HAE cultures inhibits viral infection, suggesting that KLK13 is necessary for HCoV-HKU1 infection.
Fig. 3. HCoV-HKU1 does not replicate in HAE cultures deficient in KLK13.
(A to C) Primary human epithelial cells were transduced with lentiviral vectors expressing GFP (HAE_GFP), empty pLKO.1-TRC vector (HAE_vector), or shRNA specific for KLK13 mRNA (HAE_shKLK13) and differentiated to form HAE cultures. As a control, HAE cultures differentiated from untransduced cells were used (HAE_ctrl). (A) KLK13 mRNA was evaluated before (− puro) and after puromycin (+ puro) selection of positively transduced cells. ACTB mRNA was used as an internal control. (B) Microscopic examination of all HAE cultures after 4 weeks of culture in ALI at 37°C. The images show the fully differentiated, genetically engineered HAE cultures in which KLK13 was silenced and GFP was overexpressed. Scale bars, 200 μm. (C) All HAE cultures were inoculated with HCoV-HKU1 (106 RNA copies/ml) for 2 hours at 32°C and cultured for 5 days. Each day p.i., 100 μl of PBS was applied to the apical surface of the HAE cell cultures and collected after 10 min of incubation at 32°C. Replication of HCoV-HKU1 was evaluated by RT-qPCR analysis. The data are presented as RNA copy number/ml (left) and as log removal value (LRV) compared to the untreated sample (right). The assays were performed twice, each time in triplicate (N = 3), and average values with standard errors are presented. Statistical significance was assessed with the Kruskal-Wallis test. *P < 0.05.
KLK13 enables the entry of cells by HCoV-HKU1 pseudoviruses
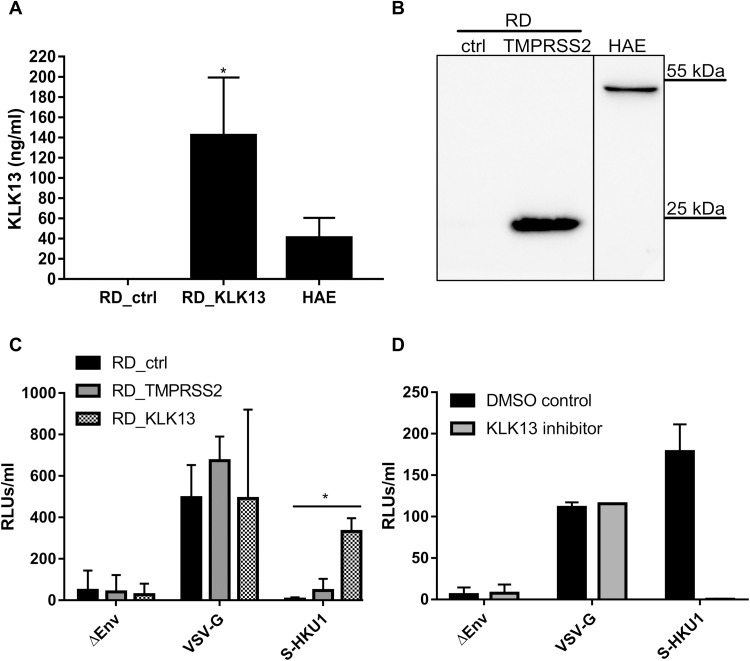
We next asked whether this enzyme was a determinant of the cell and tissue specificity of HCoV-HKU1. Previous studies showed that rhabdomyosarcoma (RD) cells support virus attachment through sialic acids but that this does not enable entry (26). To test whether cell surface proteases rendered RD cells permissive for infection, we generated RD cells expressing human KLK13 or TMPRSS2 proteases, the latter of which were used as a control. RD cells were transduced with lentiviral vectors encoding KLK13 (RD_KLK13) or TMPRSS2 (RD_TMPRSS2) or with control vector (RD_ctrl). KLK13 expression was assessed in the cell culture medium collected from RD_KLK13 and RD_ctrl cells, as well as HAE cultures with a KLK13-specific enzyme-linked immunosorbent assay (ELISA) (Fig. 4A) (65, 66). The presence of TMPRSS2 in RD_TMPRSS2 cells was confirmed by Western blotting analysis (Fig. 4B). The TMPRSS2 band in the RD cell sample migrated with an apparent molecular mass of 25 kD, which corresponds to one of the naturally occurring splice variants of the protein (Fig. 4B). Subsequently, we infected RD_ctrl, RD_KLK13, and RD_TMPRSS2 cells with HIV particles pseudotyped with HCoV-HKU1 S glycoprotein (S-HKU1) or control vesicular stomatitis virus G protein (VSV-G) or with particles lacking the fusion protein (ΔEnv). After 3 days of culture at 37°C, pseudovirus entry was quantified by measurement of luciferase activity (Fig. 4C). All cell cultures were effectively infected with control VSV-G vectors, whereas only the RD_KLK13 cells were permissive to S-HKU1 pseudoviruses. This indicates that KLK13 was involved in HCoV-HKU1 entry and that the expression of TMPRSS2 had no such effect. Furthermore, S-HKU1, VSV-G, and ΔEnv pseudoviruses were overlaid onto fully differentiated HAE cultures in the presence of KLK13 inhibitor (10 μM) or DMSO. After 3 days of culture at 37°C, pseudovirus entry was quantified by measurement of luciferase activity. Despite there being a low transduction efficiency in the HAE cultures, we observed an increase in luciferase activity in cultures infected with S-HKU1 pseudoviruses, compared to those incubated with ΔEnv pseudoviruses, which was abrogated in the presence of the KLK13 inhibitor (Fig. 4D). Overall, these data suggest that KLK13 activity mediates HCoV-HKU1 entry into cells.
Fig. 4. RD cells expressing KLK13 are permissive for HCoV-HKU1 pseudoviruses.
(A) RD cells were transduced with lentiviral vectors expressing the KLK13 gene (KLK13) or with empty vector (ctrl). The abundance of KLK13 secreted by RD cells and HAE cultures into the cell culture medium was determined with a KLK13-specific ELISA. The assays were performed twice, each time in triplicate (N = 3). Statistical significance was assessed with the Mann-Whitney test. *P < 0.05. (B) RD cells were transduced with lentiviral vectors expressing the TMPRSS2 gene (TMPRSS2) or empty vector (ctrl). After blasticidin selection, the cells were lysed and proteins were resolved by SDS-PAGE and analyzed by Western blotting. TMPRSS2 was detected in RD cell lysates (50 μg of protein per lane) and HAE cultures lysate (25 μg of protein per lane) using a specific antibody. The vertical black line indicates that the lanes are not contiguous. Blots are representative of three experiments. (C) RD control cells (RD_ctrl), TMPRSS2-expressing RD cells (RD_TMPRSS2), and KLK13-expressing RD cells (RD_KLK13) were transduced with HIV pseudoviruses decorated with VSV-G protein (VSV-G) or S-HKU1 glycoprotein (S-HKU1) or with control viruses without the fusion protein (ΔEnv). After 72 hours at 37°C, pseudovirus entry was measured by measurement of the luminescence signal in the cell lysates [relative light units (RLUs)/ml of lysate sample]. The assays were performed twice, each time in triplicate (N = 3). Data are means ± SEM. Statistical significance was assessed with the Kruskal-Wallis test. *P < 0.05. (D) HAE cultures were inoculated with HIV pseudoviruses expressing the VSV-G control protein or S-HKU1 or with control viruses without the fusion protein (ΔEnv) in the presence of KLK13 inhibitor (10 μM) or DMSO. After 72 hours at 37°C, the entry of the pseudoviruses was determined by measuring the luminescence signal in the cell lysates. The assays were performed in duplicate (N = 2). Data are means ± SEM.
KLK13 enables the replication of HCoV-HKU1 in RD cells
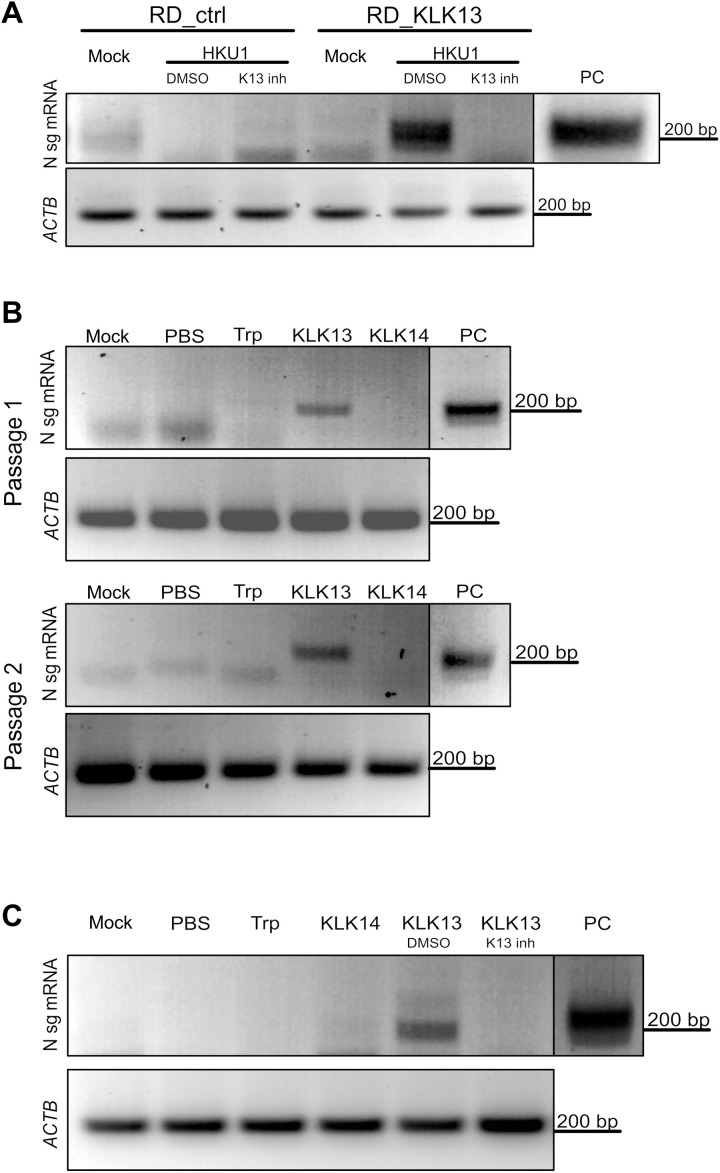
Our results thus far showed that KLK13 expression on RD cells was sufficient for the entry of a pseudotyped virus expressing the HCoV-HKU1 S protein. Next, we tested whether the presence of KLK13 rendered RD cells permissive for HCoV-HKU1 infection. Thus, we infected RD_ctrl and RD_KLK13 cells with HCoV-HKU1 (108 RNA copies/ml) and incubated the cell cultures for 7 days at 32°C in the presence or absence of a KLK13 inhibitor (10 μM) or DMSO. Cellular RNA was then isolated, and the presence of HCoV-HKU1 N subgenomic mRNA (N sg mRNA), which is considered to be a hallmark of coronaviral infection, was assessed as described previously (14). We detected HCoV-HKU1 sg mRNA in RD_KLK13 cells, but there was no detectable signal in cell cultures treated with the KLK13 inhibitor nor in RD_ctrl cells (Fig. 5A).
Fig. 5. HCoV-HKU1 replicates in RD cells expressing KLK13.
(A) Control (RD_ctrl) and KLK13-expressing RD cells (RD_KLK13) were inoculated with HCoV-HKU1 (106 RNA copies/ml) or were left uninfected (mock) in the presence of 10 μM KLK 13 inhibitor (K13 inh) or with DMSO as a control. After 7 days of culture at 32°C, total RNA was isolated and reverse-transcribed, and subgenomic mRNA for the viral N protein was detected by seminested PCR analysis. ACTB was used as an internal control. PC, positive control from virus-infected HAE cultures. (B) HCoV-HKU1 was incubated with 200 nM trypsin (Trp), KLK13, KLK14, or PBS for 2 hours at 32°C and further applied onto the RD cells. Top: After 7 days at 32°C, total RNA was isolated and reverse-transcribed, and subgenomic mRNA for the viral N protein was detected by seminested PCR analysis (passage 1). Bottom: In addition, 1 ml of the cell culture medium was harvested and applied to freshly seeded RD cells with medium supplemented with fresh enzymes. After 7 days at 32°C, subgenomic mRNA for the N protein was detected by seminested PCR (passage 2). ACTB was used as an internal control. (C) HCoV-HKU1 was incubated with 200 nM trypsin (Trp), KLK14, KLK13, KLK13 in the presence of the KLK13 inhibitor (K13 inh), DMSO (DMSO), or PBS for 2 hours at 32°C and further applied onto RD cells. Subgenomic mRNA for N protein was detected by seminested PCR. ACTB was used as an internal control. The vertical black lines indicate that the lanes are not contiguous.
To further confirm the role of KLK13 during HCoV-HKU1 infection, RD cells were supplemented with purified human KLK13 or KLK14 (61); the latter was used as a negative control. Virus stock was incubated for 2 hours at 32°C with 200 nM KLK13 or KLK14, trypsin, or control [phosphate-buffered saline (PBS)]. Next, RD cells were incubated for 7 days at 32°C with the virus [diluted 10-fold in Dulbecco’s minimal essential medium (DMEM)] or mock samples (in DMEM), after which cellular RNA was isolated, and HCoV-HKU1 infection was analyzed by means of N sg mRNA detection. Again, we found that N sg mRNA was produced only in the presence of KLK13 (Fig. 5B). Furthermore, we passaged HCoV-HKU1 twice in RD cells. Briefly, 1 ml of cell culture medium from the first experiment was transferred to fresh RD cells, and fresh enzymes were added (at a final concentration of 200 nM). Cultures were then incubated at 32°C for 7 days. Cellular RNA was isolated, and HCoV-HKU1 infection was monitored by detecting N sg mRNA. The infection occurred only in the presence of KLK13 (Fig. 5B). However, we observed no cytopathic effects, and replication levels were very low (no significant increase over control levels as assessed by RT-qPCR analysis; fig. S1). To further test the effect of KLK13 on the replication of HCoV-HKU1 in RD cells, the virus stock was incubated with purified KLK13 (200 nM) and incubated in the presence or absence of a KLK13 inhibitor (10 μM) or DMSO. After 2 hours at 32°C, pretreated virus stock was diluted in medium as described earlier and overlaid on the RD cells. After 7 days at 32°C, we evaluated the presence of the HCoV-HKU1 N sg mRNA. We found that viral replication was detected only after KLK13 treatment and that supplementation with the KLK13 inhibitor reversed this effect (Fig. 5C).
KLK13 primes the HCoV-HKU1 S protein
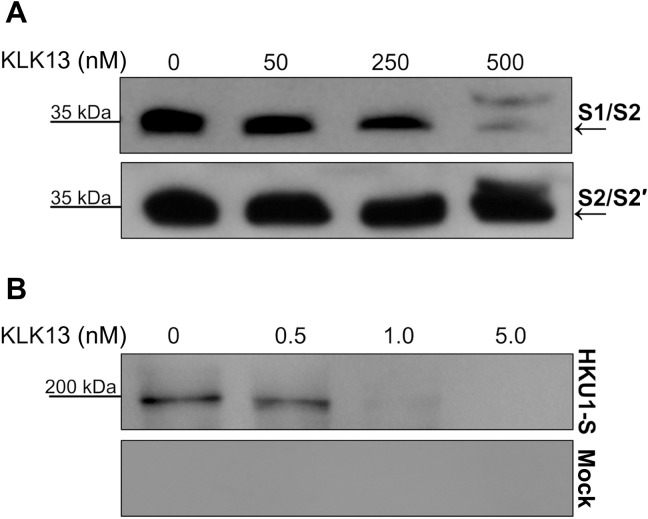
Expression of KLK13 by cells previously resistant to HCoV-HKU1 rendered them susceptible. Therefore, we asked whether this was due to proteolytic activation of the viral S protein. We tested this hypothesis using the CleavEx method, in which a peptide of interest is exposed in the N-terminal region of the proteolytically resistant HmuY carrier protein. Briefly, two-hybrid His-tagged CleavEx proteins were prepared, both harboring eight–amino acid peptide sequences of the HCoV-HKU1 S protein. The first peptide contained the S1/S2 cleavage site (amino acid residues 757 to 764), which, in some coronaviruses, is activated during protein biosynthesis, during virus exocytosis, or after receptor engagement. The second peptide harbored the S2′ cleavage site (amino acid residues 901 to 908), which is an additional region that is prone to proteolytic cleavage (25, 38). The proteins were purified and further incubated for 3 hours at 37°C with increasing concentrations of purified KLK13. Subsequently, the proteins were resolved by SDS–polyacrylamide gel electrophoresis (PAGE) and detected by Western blotting with antibodies specific for His-tagged proteins. The analysis showed that, in the presence of 500 nM KLK13, the CleavEx protein harboring the S1/S2 cleavage site was degraded; however, the CleavEx protein harboring the S2′ site remained intact (Fig. 6A). Because KLKs are produced as proforms that undergo self-activation, an additional band of His-tagged purified pro-KLK13 was observed after treatment with 500 nM KLK13 (Fig. 6A). The product of the S1/S2 cleavage was further sequenced by N-terminal Edman degradation showing the following sequence: R↓SISA, which corresponds to the S1/S2 site (file S1). This result shows that the S1/S2 region of the HCoV-HKU1 S protein is prone to KLK13-mediated cleavage.
Fig. 6. KLK13 cleaves the HCoV-HKU1 S protein between the S1 and S2 domains.
(A) Fifteen nanograms of CleavEx proteins harboring the S1/S2 or S2′ sites was incubated at 37°C for 3 hours with the indicated concentrations of purified KLK13 protein. The samples were then denatured at 95°C, and the proteins were resolved by SDS-PAGE and analyzed by Western blotting with antibodies specific for His-tagged proteins. Blots are representative of three experiments. (B) The full-length HKU1-S protein or control sample was incubated at 37°C for 3 hours with the indicated concentrations of purified KLK13 protein. The samples were then denatured at 95°C, and the proteins were resolved by SDS-PAGE and analyzed by Western blotting with HRP-conjugated antibody against the His tag to detect the full-length protein with the N-terminal His tag (~200 kD) and the S1 after KLK13 cleavage (~80 kD). Blots are representative of three experiments.
Furthermore, we aimed to confirm the cleavage using a full-length S protein of HCoV-HKU1 (HKU1-S). For this, we expressed the HKU1-S in human embryonic kidney (HEK) 293T cells, purified the protein using 6× His tag, and incubated it for 3 hours at 37°C with increasing concentrations of purified KLK13. Subsequently, HKU-S or control sample was resolved by SDS-PAGE and detected by Western blotting with antibodies specific to the tag. The analysis showed that, in the presence of 1 μM KLK13, the HKU1-S was degraded (Fig. 6B). The full-length S protein was observed at ~200 kD, which is consistent with previous report (1). After KLK13 treatment, the S protein band diminished, and the band corresponding to the S1 domain (~80 kD) became visible (Fig. 6B).
DISCUSSION
Receptor recognition is the first essential step of the viral infection process. The coronaviral S protein mediates virus entry into host cells by binding to a specific receptor. A combination of stimuli, for example, receptor binding, proteolytic cleavage, and exposure to low pH, results in rearrangement of the S protein and, consequently, membrane fusion and viral entry (1). Although the structures of both the HCoV-HKU1 S ectodomain and the receptor-binding domain have been resolved, the receptor determinant remains unknown (28, 38, 67). Previously, we showed that HCoV-HKU1 uses O-acetylated sialic acids on host cells as an attachment receptor (26). Here, we present data demonstrating that the protease KLK13 is required for HCoV-HKU1 infection of fully differentiated respiratory epithelial cells reconstituting the respiratory epithelium ex vivo.
Human KLKs mediate multiple physiological processes, including skin desquamation, tooth enamel formation, kidney and brain function, and synaptic neural plasticity (68–77). In addition, studies have demonstrated a role for some KLKs during viral infections. For example, KLK8 plays a role in the proteolytic activation of the human papillomavirus capsid protein, thereby mediating virus entry into host cells (78). KLK5 and KLK12 are secreted into the respiratory tract, where they support replication of the influenza A virus by cleaving the hemagglutinin protein (79, 80); however, these proteins belong to a large pool of cell surface proteases, the orchestrated action of which promotes virus replication.
The biological activities of KLKs include processing of bradykinins, which have been discussed in the context of SARS-CoV-2. Available reports indicate the importance of RAS, Ace-ACE2 balance, and bradykinin activity during coronavirus disease 2019. The involvement of KLK-related proteases in coronaviral infection may therefore reveal previously uncharacterized mechanisms of pathogenesis within the airway epithelial tissue. However, note that this activity is limited to plasma KLKs and not tissue-associated enzymes such as KLK13 (81, 82).
Here, we found that the yield of HCoV-HKU1 from HAE cultures was reduced in the presence of SPINK6 (an inhibitor of KLK13) (62, 63) and camostat (a broad range inhibitor of serine proteases). However, the first compound also inhibits other KLKs (63), whereas the second blocks the activity of a wide range of serine proteases and was used previously to demonstrate the role of TMPRSS2/4 proteases during viral infection (34, 45, 51, 64, 83, 84). The relatively low-level inhibition of HCoV-HKU1 replication in the presence of SPINK6 possibly results from non-optimal compound concentration and cytotoxicity at higher concentrations (63). Note that even the low level of inhibition observed was sufficient to limit viral replication. SPINK6 was identified previously as KLK family-specific, fully reversible inhibitor with a Ki in the nanomolar range. This broad substrate specificity may result in greater toxicity, but, at the same time, the presence of other proteases will affect the availability and stability of SPINK6. To overcome these limitations, we optimized KLK13 inhibition with a small-molecule, reversible inhibitor selective against KLK13. Here, we observed inhibition similar to that observed for camostat. We believe that high activity of KLK13-specific inhibitor, in comparison to the broadly selective one, supports our conclusion that KLK13 is an essential protease in HCoV-HKU1 infection.
Broad-spectrum protease inhibitors are now used widely for virus research, although their nonspecific activity makes the results obtained through their use equivocal. For example, Matsuyama et al. (85) showed that the furin inhibitor dec-RVKR-CMK interferes with the activity of several proteases and that its previously described inhibitory activity during MERS-CoV infection is not specific to furin; instead, its activity is due to nonspecific inhibition of cathepsin L and TMPRSS2. We tried to use specific KLK inhibitors developed in our laboratory (61). Considering the small arsenal of tools available to researchers studying KLKs, only three compounds were readily available. Treating HAE cultures with these inhibitors revealed that only compounds designed to inhibit KLK13 hampered HCoV-HKU1 replication. However, the high similarity between different KLKs makes one doubt the specificity of these inhibitors, despite their performance in biochemical assays. Therefore, we decided to silence KLK13 in HAE cultures. This abolished virus replication in vitro, thereby confirming the importance of KLK13 during infection. KLK13 is thought to be secreted and membrane bound because of its ability to bind to surface glycosaminoglycans (86, 87).
In the current study, we show that HCoV-HKU1 infection in HAE cultures modulates the expression of different KLKs, including KLK13. In our study, KLK expression was tested by semiquantitative PCR, and for that reason, we were unable to show the extent of KLK modulation after HCoV-HKU1 infection. However, the pattern of virus-induced expression of several KLKs could be observed. The mechanism of activation of KLKs is a complex process, and until now, it has only been proven that most KLK-encoding genes are regulated by steroids and other hormones (88). Note that KLK expression is regulated in a similar manner, and the induction of a single gene usually results in overexpression of the whole cluster (89, 90). Although one may assume that the virus stimulates KLK13 production to promote the infection, this increase in KLK13 abundance is likely a natural response of the damaged tissue, because KLKs also participate in tissue regeneration (91–93). Furthermore, the increased expression of KLKs may be a response to the inflammatory process, because Seliga et al. (94) demonstrated that the KLK-kinin system is a potent modulator of innate immune responses.
The experiments performed here showed the importance of KLK13 for viral entry into susceptible cells; therefore, we speculated that the scattered distribution of different KLKs in different tissues may be one of the determinants of HCoV-HKU1 tropism (53, 95). We tested the purified enzyme expressed in the eukaryotic cells; however, we also developed a cell line constitutively expressing the enzyme. As an in vitro model for our studies, we used RD cells previously reported to have attachment receptors for the virus (26). Here, using pseudoviruses decorated with S-HKU1 proteins, we showed that the presence of KLK13 on RD cells was sufficient for viral entry and rendered these cells permissive for infection. We believe that during HCoV-HKU1 infection in RD cells, the S protein may be alternatively activated by cellular cathepsins during viral endocytosis, as was shown for other coronaviruses (44, 96, 97). Furthermore, we observed that RD cells supported viral replication in the presence of KLK13 and that this effect was reversed in the presence of the specific KLK13 inhibitor. We were, however, unable to culture the virus to high yields. HCoV-HKU1 replication in KLK13-expressing RD cells remained inefficient, and RT-qPCR–based assessment did not reveal significant increases in the amounts of viral RNA. For that reason, we are only able to detect viral sg mRNAs, which are considered to be the hallmark of coronaviral replication. We believe that this may be due to suboptimal infection conditions, which may include inappropriate KLK13 concentrations or low density of the entry receptor. In addition, it is possible that RD cells may not support efficient replication of the virus due to factors unrelated to the entry process. Nonetheless, our results indicate that the HCoV-HKU1 entry receptor is present on RD cells, and we were able to trigger viral entry and replication. Thus, these findings warrant further exploration.
Most coronaviral S proteins are processed into S1 and S2 subunits by host proteases, mainly furin or furin-like protease and TMPRSS2 or TMPRSS2-like proteases, which enables conformational changes in the S protein and leads to fusion of the viral and cellular membranes (1, 98). As shown in the study by Kirchdoerfer et al. (38), the HCoV-HKU1 S protein has two regions that are prone to proteolytic activation: the S1/S2 furin cleavage site and a secondary cleavage site termed S2′, which is adjacent to a potential fusion peptide. Whereas the S1/S2 site is believed to be processed by furin during protein biosynthesis, the S2′ site is expected to be cleaved during viral entry by the TMPRSS2-like protease. These structural data have not been confirmed using an infectious virus. Because we already knew that KLK13 was sufficient for HCoV-HKU1 infection of naturally nonpermissive RD cells, we aimed to investigate whether this was the direct result of KLK13-mediated proteolytic cleavage of the S protein. For this, we used the CleavEx method, in which a peptide of interest is coupled to the carrier HmuY protein and then undergoes proteolytic cleavage by the tested enzyme. We found that the S1/S2 site was efficiently cleaved by KLK13, whereas the S2′ region remained intact. Because the CleavEx technique is a convenient surrogate system that enables precise mapping of the cleavage site, it has some limitations. To ensure the reliability of our experiments, purified full-length HCoV-HKU1 S protein was subjected to proteolytic cleavage. In this experiment, we also observed efficient cleavage of the HCoV-HKU1 S protein by KLK13. KLK13 shares specificity with furin and furin-like proteases (56, 61, 63), and our results complement previous data on the presence of the furin-cleavage site in the S1/S2 region of the HCoV-HKU1 S. However, whereas for the purified protein or pseudovirus particles, the S protein was processed during the protein maturation, and for the infectious virus, extracellular KLK seemed to be essential. One may link this observation with the fact that virus release follows a different route than that of a single protein and that coronaviruses bud from the endoplasmic-reticulum–Golgi intermediate compartment, whereas lentiviruses are believed to bud from the plasma membrane.
Although the results presented here suggest that KLK13 can process the HCoV-HKU1 S protein, one may question whether this cleavage event is sufficient for HCoV-HKU1 entry. In the case of MERS-CoV, two consecutive enzymatic scissions are required for activation of the S protein. In this scenario, KLK13 might prime the HCoV-HKU1 S at the S1/S2 site, enabling scission at the S2′ site by the TMPRSS2 or another host protease (38, 40, 99). This may be one of the factors limiting the replication of HCoV-HKU1 in RD_KLK13 cells, as only minimal replication was observable in our hands.
In conclusion, we showed data suggesting that KLK13 is a key determinant of HCoV-HKU1 tropism. This finding may explain why, since its first identification in 2004, all efforts to culture HCoV-HKU1 in standard cell lines have failed. We believe that this study increases our knowledge of HCoV-HKU1 and may promote the future in-depth investigation of coronaviruses. Considering the increasing number and diversity of coronaviruses, and the proven propensity of coronaviruses to cross the species barrier and cause severe diseases in humans, further research on this group of pathogens is necessary.
MATERIALS AND METHODS
Plasmid constructs
The sequences encoding KLK13 and TMPRSS2 were amplified by PCR using cDNA obtained from HAE cultures. Each PCR product was subcloned into the pWPI plasmid for lentivirus production, and sequences were verified. The pLKO.1-TRC cloning vector was a gift from D. Root (Addgene plasmid no. 10878) (100). Oligonucleotides for the generation of shRNA against KLK13 (three different shRNAs targeting the exons encoding the active site) were hybridized and subcloned into pLKO.1-TRC vector. The full-length HKU1-S sequence was amplified by PCR using pCAGGS/HKU1-S plasmid, which was a gift from X. Huang (National Institute of Biological Sciences, Changping, Beijing, China). The PCR product was subcloned into pSecTag2 cloning vector, and its sequence was verified. Primer sequences are provided in table S2.
Cell culture
RD [Homo sapiens muscle rhabdomyosarcoma; American Type Culture Collection (ATCC): CCL-135] and 293T (H. sapiens kidney epithelial; ATCC: CRL-3216) cells were cultured in DMEM (Thermo Fisher Scientific, Poland) supplemented with 3% heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific, Poland) and antibiotics: penicillin (100 U/ml), streptomycin (100 μg/ml), and ciprofloxacin (5 μg/ml). Cells were maintained at 37°C under 5% CO2.
HAE cultures
Human airway epithelial cells were isolated from conductive airways resected from transplant patients. The study was approved by the Bioethical Committee of the Medical University of Silesia in Katowice, Poland (approval no: KNW/0022/KB1/17/10 dated 16 February 2010). Written consent was obtained from all patients. Cells were dislodged by protease treatment and later were mechanically detached from the connective tissue. The resulting primary cells were first cultured in selective medium to proliferate in the presence of the Rho-associated protein kinase inhibitor (Y-27632, 10 μg/ml, Sigma-Aldrich, Poland), which enhances cell proliferation and enables the serial passaging of airway epithelial cells without compromising their phenotype (101). The cells were then trypsinized and transferred onto permeable Transwell insert supports (6.5-mm diameter). Cell differentiation was stimulated by the media additives, and removal of media from the apical side after the cells reached confluence. Cells were cultured for 4 to 6 weeks to form well-differentiated, pseudostratified, and mucociliary epithelium (HAE cultures) (102). All experiments were performed in accordance with the relevant guidelines and regulations.
Cell viability assay
HAE cultures were prepared as described earlier. Cell viability was assessed with the XTT Cell Viability Assay (Biological Industries, Israel), according to the manufacturer’s instructions. Briefly, on the day of the assay, 100 μl of 1× PBS containing 30 μl of the activated XTT solution was added to each well per culture insert. After incubation for 2 hours at 37°C, the solution was transferred onto a 96-well plate, and the color change was assessed with a colorimeter (Spectra MAX 250, Molecular Devices) at a wavelength of 490 nm. The obtained results were further normalized to the control sample, where cell viability was set to 100%.
Virus infection
HAE cultures were washed thrice with 100 μl of PBS, after which they were inoculated with HCoV-HKU1 (strain Caen 1) or were subjected to mock inoculation (cell lysate). After a 2-hour incubation at 32°C, unbound virions were removed by washing with 100 μl of PBS, and the HAE cultures were maintained at an ALI until the end of the experiment. Because of the lack of a permissive cell line, it was not possible to titrate the virus stock for infection experiments, and the inoculum was quantified by RT-qPCR analysis. RD cells grown to 90% confluency were infected with HCoV-HKU1 (108 RNA copies/ml) in DMEM supplemented with 3% heat-inactivated FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were incubated for 7 days at 32°C, washed thrice with PBS, and collected for RNA isolation in Fenozol reagent (A&A Biotechnology, Poland). All research involving infectious material was performed while adhering to biosafety regulations. All research involving genetic modifications was performed in compliance with national and international regulations.
Lentivirus production and transduction
The 293T cells were seeded on 10-cm2 dishes, cultured for 24 hours at 37°C with 5% CO2, and transfected with psPAX, pMD2G, and third transfer plasmid (pWPI/KLK13, pLKO.1-TRC/shrnaKLK13, or Lego-G2) with polyethyleneimine (Sigma-Aldrich, Poland). Both psPAX (Addgene plasmid no. 12260) and pMD2G (Addgene plasmid no. 12259) were gifts from D. Trono. The pLKO.1-TRC cloning vector was a gift from D. Root (Addgene plasmid no. 10878) (100). The cells were further cultured for 96 hours at 37°C with 5% CO2, and lentiviral particles were collected every 24 hours and stored at 4°C. Lentivirus stocks were concentrated 25-fold with centrifugal protein concentrators (Amicon Ultra, 10-kDa cutoff; Merck, Poland) and stored at −80°C. RD cells were seeded in T75 flasks, cultured for 24 hours at 37°C with 5% CO2, and transduced with lentiviral particles expressing KLK13 or TMPRSS2 or with control vector in the presence of polybrene (4 μg/ml; Sigma-Aldrich, Poland). Cells were further cultured for 72 hours at 37°C with 5% CO2, and positively transduced cells were selected with blasticidin (2 μg/ml; Sigma-Aldrich, Poland). Primary human epithelial cells seeded on 10-cm2 dishes were cultured in bronchial epithelial cell growth medium and transduced with lentiviral particles expressing KLK13-specific shRNAs (a set of three) or GFP in the presence of polybrene (5 μg/ml; Sigma-Aldrich, Poland). Cells were further cultured for 72 hours at 37°C with 5% CO2, and the positively transduced cells were selected with puromycin (5 μg/ml; Sigma-Aldrich, Poland). Selected cells were plated on insert supports and further cultured in an ALI in the presence of puromycin (1 μg/ml).
Pseudoviruses
The 293T cells were seeded onto six-well plates, cultured for 24 hours at 37°C with 5% CO2, and transfected with polyethyleneimine (Sigma-Aldrich, Poland) with the lentiviral packaging plasmid (psPAX), the VSV-G envelope plasmid (pMD2G) or HCoV-HKU1 S glycoprotein (pCAGGS-HKU1-S), and a third plasmid encoding luciferase (pRR Luciferase). pRR Luciferase was a gift from P. Khavari (Addgene plasmid no. 120798) (103). Cells were cultured for a further 72 hours at 37°C with 5% CO2, and pseudoviruses were collected every 24 hours and stored at 4°C. RD cells were seeded in 48-well plates, cultured for 24 hours at 37°C with 5% CO2, and transduced with pseudoviruses expressing VSV-G or S-HKU1 proteins or lacking the fusion protein (ΔEnv) in the presence of polybrene (4 μg/ml). HAE cell cultures were washed thrice with 100 μl of PBS and subsequently inoculated with S-HKU1 or VSV-G pseudoviruses. After 4 hours of incubation at 37°C, unbound virions were removed by washing with 100 μl of PBS, and the HAE cultures were cultured at an ALI. The cells were further cultured for 72 hours at 37°C with 5% CO2 and lysed in luciferase substrate buffer (Bright-Glo; Promega, Poland). Lysates were transferred onto white 96-well plates, and luciferase activity was measured on a microplate reader Gemini EM (Molecular Devices, UK).
Isolation of nucleic acids and reverse transcription
A viral DNA/RNA Kit (A&A Biotechnology, Poland) was used for nucleic acid isolation from cell culture medium, according to the manufacturer’s instructions. Cellular RNA was isolated with Fenozol reagent (A&A Biotechnology, Poland), followed by treatment with deoxyribonuclease I (DNase I) (Thermo Fisher Scientific, Poland). cDNA samples were prepared with a High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Poland), according to the manufacturer’s instructions.
Polymerase chain reaction
Human KLK mRNAs were reverse-transcribed and amplified in a reaction mixture containing 1× Dream Taq Green PCR master mix (Thermo Fisher Scientific, Poland) and appropriate primers (500 nM each, table S3). Primers specific for KLK1, KLK2, and KLKs 4 to 15 were described previously (104). Primers specific for KLK3 were developed and optimized in our laboratory. ACTB was used as a housekeeping gene reference. The reaction was performed as follows: 5 min at 95°C, followed by 35 cycles of (30 s at 95°C, 20 s at 59°C, and 20 s at 72°C), followed by 10 min at 72°C.
Quantitative PCR
The HCoV-HKU1 RNA yield was assessed by RT-qPCR analysis (7500 Fast Real-Time PCR; Life Technologies, Poland). The cDNA was amplified in a reaction mixture containing 1× TaqMan Universal PCR Master Mix (Thermo Fisher Scientific, Poland), in the presence of FAM/TAMRA (6-carboxyfluorescein/6-carboxytetramethylrhodamine) probe (100 nM; 5′-TTG AAG GCT CAG GAA GGT CTG CTT CTA A-3′) and primers [450 nM each; 5′-CTG GTA CGA TTT TGC CTC AA-3′ (forward) and 5′-ATT ATT GGG TCC ACG TGA TTG-3′ (reverse)] (105). The reaction was performed as follows: 2 min at 50°C and 10 min at 92°C, followed by 40 cycles of (15 s at 92°C and 1 min at 60°C).
Detection of HCoV-HKU1 N sg mRNA
Total nucleic acids were isolated from virus- or mock-infected cells at 7 days p.i. with Fenozol reagent (A&A Biotechnology, Poland) according to the manufacturer’s instructions. Reverse transcription was performed with a high-capacity cDNA reverse transcription kit (Life Technologies, Poland) according to the manufacturer’s instructions. Viral cDNA was amplified in a 20-μl reaction mixture containing 1× Dream Taq Green PCR master mix (Thermo Fisher Scientific, Poland) and primers (500 nM each). The following primers were used to amplify HCoV-HKU1 sg mRNA: common sense primer (leader sequence), 5′-TCT TGT CAG ATC TCA TTA AAT CTA AAC T-3′; nucleocapsid antisense for the first PCR, 5′-AAC TCC TTG ACC ATC TGA AAA TTT-3′; nucleocapsid antisense for the nested PCR, 5′-AGG AAT AAT GTG GGA TAG TAT TT-3′. The conditions were as follows: 3 min at 95°C, 35 cycles (30 cycles for nested PCR) of (30 s at 95°C, 30 s at 49°C, and 20 s at 72°C), which was followed by 5 min at 72°C and 10 min at 4°C. The PCR products were resolved on 1% agarose gels [tris-acetate-EDTA (TAE) buffer] and analyzed with molecular imaging software (Thermo Fisher Scientific, Poland).
Detection of TMPRSS2
After they underwent selection in blasticidin-containing medium, RD cells expressing TMPRSS2 (RD_TMPRSS2) or KLK13 (RD_KLK13) or control cells (RD_ctrl) were scraped and collected by centrifugation. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer [50 mM tris, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS (pH 7.5)], boiled for 5 min, cooled on ice, and resolved on a 10% polyacrylamide gel alongside dual-color Page Ruler Prestained Protein size markers (Thermo Fisher Scientific, Poland). The separated proteins were then transferred onto a Westran S polyvinylidene difluoride (PVDF) membrane (GE Healthcare, Poland) by wet blotting (Bio-Rad, Poland) for 1 hour at 100 V in transfer buffer (25 mM tris, 192 mM glycine, and 20% methanol) at 4°C. The membranes were blocked by overnight incubation at 4°C in Tris-buffered saline (TBS)–Tween (0.1%) buffer supplemented with 5% skimmed milk (BioShop, Canada). The blots were incubated with a mouse monoclonal anti–TMPRSS2 antibody (clone P5H9-A3; 1:500 dilution; Sigma-Aldrich, Poland), followed by incubation with a horseradish peroxidase (HRP)–labeled anti-mouse immunoglobulin G (65 ng/ml; Dako, Denmark) diluted in 5% skimmed milk/TBS-Tween (0.1%). The signal was developed with the Pierce ECL Western blotting substrate (Thermo Fisher Scientific, Poland) and visualized with the ChemiDoc Imaging System (Bio-Rad, Poland).
KLK13 ELISA
Nunc U16 Maxisorp ELISA strips (Thermo Fisher Scientific, Poland) were coated overnight with 100 μl of anti-human KLK13(33-1) antibodies [5 μg/ml in 50 mM tris (pH 7.8)]. Twenty-four hours later, the wells were washed three times with washing buffer [5 mM tris (pH 7.8), 150 mM NaCl, and 0.05% Tween] and blocked for 2 hours with 200 μl of blocking solution [6% bovine serum albumin (BSA) in 50 mM tris (pH 7.8)]. Standards (recombinant human KLK13, 100 μl), controls, and samples diluted in assay buffer [50 mM tris (pH 7.8), 0.05% Tween, and 6% BSA] were applied to the wells and incubated for 2 hours at room temperature with gentle shaking. Wells were washed four times with washing buffer. Detection antibodies and biotinylated anti–human KLK13(27-1) antibodies (100 μl at 0.25 μg/ml) in assay buffer were applied and incubated for 1 hour at room temperature. Wells were washed four times with washing buffer. Streptavidin-HRP conjugate (100 μl at 0.05 μg/ml; Jackson Immuno Research, USA) in assay diluent was added and incubated for 20 min at room temperature. Wells were washed six times with washing buffer. 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate solution (100 μl, Becton Dickinson, Poland) was added, and the plate was incubated until color development was complete. 2 N H2SO4 (50 μl) was added to each well to stop the reaction. The absorbance was measured at 450 nm (with correction at 570 nm) on a microplate reader (Tecan, Switzerland). Each sample was analyzed in triplicate.
Expression and purification of KLK13 and KLK14
The sequence of the proKLK13-encoding gene was amplified from cDNA obtained from HAE cultures with specific primers. The codon-optimized gene encoding proKLK14 was custom-synthesized (Thermo Fisher Scientific, Poland). The products were subcloned into the pLEXSY_I-blecherry3 plasmid (Jena Bioscience, Germany), and the resulting constructs were verified by sequencing. All preparations for transfection, selection, and expression of the host Leishmania tarentolae strain T7-TR were performed according to the Jena Bioscience protocol for inducible expression of recombinant proteins secreted to medium (LEXSinduce Expression kit, Jena Bioscience, Germany). Expression of proKLK13 and proKLK14 was induced with 15 μg/ml of tetracycline (BioShop, Canada) and was performed for three consecutive days. Culture medium was collected, precipitated with 80% ammonium sulfate, and centrifuged at 15,000g for 30 min at 4°C. Pellets were suspended in 10 mM sodium phosphate (pH 7.5) and dialyzed overnight at 4°C into 10 mM sodium phosphate (pH 7.5). The KLKs were isolated with 6× His tag using nickel resin (GE Healthcare, Poland) according to the manufacturer’s protocol. Obtained fractions were analyzed by SDS-PAGE under reducing conditions, and fractions containing proKLK13 or proKLK14 were concentrated with Vivaspin 2 (Sartorius, Germany) and further purified by size-exclusion chromatography (Superdex s75 pg; GE Healthcare, Poland). Fractions containing proKLK13 or proKLK14 were concentrated, and the buffer was changed to 50 mM tris (pH 7.5) and 150 mM NaCl. After purification and self-activation at 37°C for 24 hours, the activities of the proteases were assessed by titration with the serine protease inhibitor Kazal-type 6 (SPINK6), as described previously (63).
Cloning of HmuY-based CleavEx fusion proteins
The fusion constructs were based on positions 26 to 216 of the Porphyromonas gingivalis HmuY protein–encoding gene (accession number ABL74281.1), which was used as a carrier protein. The sequence was amplified with Phusion DNA polymerase (Thermo Fisher Scientific, Poland) and specific primers (forward: 5′-ATA TGC GGC CGC AGA CGA GCC GAA CCA ACC CTC CA-3′; reverse: 5′-ATA CTC GAG TTA TTT AAC GGG GTA TGT ATA AGC GAA AGT GA-3′) from whole-genomic DNA isolated from P. gingivalis strain W83. PCR conditions were as follows: 98°C for 30 s, followed by denaturation at 98°C for 10 s, annealing at 68°C for 40 s, and extension at 72°C for 30 s/kb over 35 cycles with a final extension at 72°C for 7 min. The hmuY′ sequence from P. gingivalis bacteria was further amplified in three consecutive PCR reactions with primers specific to the 5′ hmuY’ fragment and 3′-specific primer introducing additional nucleotides dependent on the designed sequence (proKLK13 primers, table S4). The reaction product was ligated into a modified pETDuet plasmid (potential tryptic cleavage sites were removed from the multicloning site by Quick Change mutagenesis with Phusion DNA polymerase (Thermo Fisher Scientific, Poland). Alternatively, the designed fusion protein–encoding sequences were produced by Phusion Site-Directed Mutagenesis (Thermo Fisher Scientific, Poland) through sequence exchange of the previously prepared CleavEx construct (HKU1-S primers, table S4). The final product was transformed into competent Escherichia coli T10 cells and further purified and sequenced.
Expression and purification of CleavEx fusion proteins
Protein expression was performed in E. coli BL21 cells and was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside to the bacterial culture [when at an OD600 (optical density at 600 nm) of 0.5 to 0.6], followed by shaking for 3 hours at 37°C. The bacteria were then spun down, and the pellet was suspended in buffer A [10 mM sodium phosphate, 500 mM NaCl, and 5 mM imidazole (pH 7.4)]. The pellet suspension was then sonicated and spun down. Soluble proteins were purified with a HisTrap Excel column (GE Healthcare, Poland) in buffer A with a linear gradient of 0 to 100% of 1 M imidazole in buffer A in 20 column volumes. Fractions containing the protein of interest were pooled, and the buffer was exchanged to 50 mM tris (pH 7.5). Last, the protein of interest was purified by ion-exchange chromatography with a MonoQ 4.6/100 PE column (GE Healthcare, Poland) with a linear gradient of 0 to 100% of 50 mM tris (pH 7.5) and 1 M NaCl in 15 column volumes.
Expression and purification of the HCoV-HKU1 S protein
The 293T cells were seeded onto 60-cm2 dishes, cultured for 24 hours at 37°C with 5% CO2, and transfected with 25 μg of pSecTag2-HKU-S plasmid per dish with polyethyleneimine (Sigma-Aldrich, Poland). Cells were cultured for a further 72 hours at 37°C with 5% CO2 and collected for HKU1-S purification. Cell pellets were lysed in RIPA buffer [50 mM tris, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS (pH 7.5)] in the presence of Viscolase (1250 U/ml; A&A Biotechnology, Poland), clarified by centrifugation, and filtered [0.45-μm syringe polyethersulfone (PES) filter]. The supernatant containing 6× His-tagged S protein was mixed in a 1:2 ratio with binding buffer [20 mM NaH2PO4, 500 mM NaCl, and 20 mM imidazole (pH 7.4)] and purified using a fast performance liquid chromatography system (AKTA, GE Healthcare, Poland) with a Ni2+ HiTrap Immobilized Metal ion Affinity Chromatography (IMAC) (2 × 1 ml) column (GE Healthcare, Poland) preequilibrated with the binding buffer. The 6× His-tagged S protein was eluted with elution buffer [20 mM NaH2PO4, 500 mM NaCl, and 500 mM imidazole (pH 6.9)]. The control sample from mock-transfected cells was prepared in the same manner. Fractions containing 6× His-tagged S protein or the respective fractions from the control purification were pooled and dialyzed against PBS, 5% glycerol.
CleavEx screening assay and HKU1-S cleavage
A total of 15 ng of each CleavEx protein was incubated with 50, 250, and 500 nM KLK13 in 50 mM tris (pH 7.5). For the full-length S protein, fractions containing purified HKU1-S or mock samples were incubated with 0.5, 1.0, or 5.0 μM KLK13 in 50 mM tris (pH 7.5). Samples were incubated at 37°C for 3 hours, and the reactions were stopped with the addition of 50 mM dithiothreitol (DTT)–supplemented SDS sample buffer (1:1), boiled for 5 min, cooled on ice, and resolved on 10% SDS-PAGE gels together with dual-color Page Ruler Prestained Protein size markers (Thermo Fisher Scientific, Poland). The separated proteins were then transferred onto a Westran S PVDF membrane (GE Healthcare, Poland) by wet blotting (Bio-Rad, Poland) for 1 hour at 100 V in transfer buffer (25 mM tris, 192 mM glycine, and 20% methanol) at 4°C. The membranes were then blocked by overnight incubation at 4°C in TBS-Tween (0.1%) buffer supplemented with 5% skimmed milk (BioShop, Canada). An HRP-labeled anti–His tag antibody (1:25,000 dilution; Sigma-Aldrich, Poland) diluted in 5% skimmed milk/TBS-Tween (0.1%) was used to detect the His-tagged HmuY proteins. The signal was developed with the Pierce ECL Western blotting substrate (Thermo Fisher Scientific, Poland), and bands were visualized with the ChemiDoc Imaging System (Bio-Rad, Poland).
Identification of the cleavage site
A total of 10 μg of S1/S2 CleavEx protein was incubated with 500 nM KLK13 at 37°C for 5 hours. The reaction was stopped by the addition of 50 mM DTT-supplemented SDS sample buffer (1:1), and samples were immediately boiled for 5 min. The samples were then resolved on 10% SDS-PAGE gel together dual-color Page Ruler Prestained Protein size markers (Thermo Fisher Scientific, Poland) in the Tris-Tricine SDS-PAGE system. The separated proteins were then electrotransferred onto a Western S PVDF membrane with the Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad, Poland). The transfer was performed for 30 min at 15 V in transfer buffer [10 mM N-cyclohexyl-3-aminopropanesulfonic acid and 10% methanol (pH 11)]. The membranes were then stained with 0.025% (w/v) Coomassie Brilliant Blue R-250 (BioShop, Poland), and proteins in the bands of interest were sequenced by Edman degradation with a PPSQ-31A automatic protein sequencer (Shimadzu, Japan).
Acknowledgments
We are grateful to X. Huang for providing the pCAGGS/HKU1-S plasmid, A. Saraccino for providing the pWPI vector, D. Root for providing the pLKO.1-TRC cloning vector, L. Sasiadek for providing the KLK primers used in the study, and G. Dubin for providing reference samples. Funding: This work was supported by grants from the National Science Center UMO-2013/08/W/NZ1/00696 (J.P.), UMO-2016/22/E/NZ5/00332 (T.K.), UMO-2013/08/S/NZ6/00730 (A.N.), and UMO-2012/07/E/NZ6/01712 and UMO-2017/27/B/NZ6/02488 (K.P.). The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a partner of the Leading National Research Center supported by the Ministry of Science and Higher Education of the Republic of Poland. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. Author contributions: A.M., K.F., E.B., M.K., A.N., and P.M. conducted the experiments. A.L., T.K., M.O., M.U., and J.P. provided the materials and methods for the study. A.M. and K.P. designed the study and experiments, analyzed the data, and wrote the manuscript. K.P. and T.K. supervised the study. All authors reviewed the manuscript and approved the submitted version. All authors agreed to be personally accountable for their own contributions and to ensure that questions related to the accuracy or integrity of any part of the work are appropriately investigated, resolved, and their resolution documented in the literature. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
stke.sciencemag.org/cgi/content/full/13/659/eaba9902/DC1
Fig. S1. Supplementation of RD cells with proteases does not result in a significant change in virus yield.
Table S1. KLK inhibitors used in the study.
Table S2. Primers used for the generation of plasmid constructs.
Table S3. Primers used for the PCR-based amplification of each KLK-encoding gene.
Table S4. Primers used in the CleavEx design.
File S1. Protein sequence data.
REFERENCES AND NOTES
- 1.B. N. Fields, D. M. Knipe, P. M. Howley, Fields Virology (Wolters Kluwer Health/Lippincott Williams & Wilkins, ed. 6, 2013). [Google Scholar]
- 2.Peiris J. S., Yuen K. Y., Osterhaus A. D. M. E., Stöhr K., The severe acute respiratory syndrome. N. Engl. J. Med. 349, 2431–2441 (2003). [DOI] [PubMed] [Google Scholar]
- 3.de Groot R. J., Baker S. C., Baric R. S., Brown C. S., Drosten C., Enjuanes L., Fouchier R. A. M., Galiano M., Gorbalenya A. E., Memish Z. A., Perlman S., Poon L. L. M., Snijder E. J., Stephens G. M., Woo P. C. Y., Zaki A. M., Zambon M., Ziebuhr J., Middle East respiratory syndrome coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J. Virol. 87, 7790–7792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaki A. M., van Boheemen S., Bestebroer T. M., Osterhaus A. D. M. E., Fouchier R. A., Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012). [DOI] [PubMed] [Google Scholar]
- 5.van der Hoek L., Pyrc K., Jebbink M. F., Vermeulen-Oost W., Berkhout R. J. M., Wolthers K. C., Wertheim-van Dillen P. M. E., Kaandorp J., Spaargaren J., Berkhout B., Identification of a new human coronavirus. Nat. Med. 10, 368–373 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Hoek L., Sure K., Ihorst G., Stang A., Pyrc K., Jebbink M. F., Petersen G., Forster J., Berkhout B., Uberla K., Croup is associated with the novel coronavirus NL63. PLOS Med. 2, e240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeager C. L., Ashmun R. A., Williams R. K., Cardellichio C. B., Shapiro L. H., Look A. T., Holmes K. V., Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357, 420–422 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dijkman R., Jebbink M. F., Deijs M., Milewska A., Pyrc K., Buelow E., van der Bijl A., van der Hoek L., Replication-dependent downregulation of cellular angiotensin-converting enzyme 2 protein expression by human coronavirus NL63. J. Gen. Virol. 93, 1924–1929 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pöhlmann S., Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U.S.A. 102, 7988–7993 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pöhlmann S., Gramberg T., Wegele A., Pyrc K., van der Hoek L., Berkhout B., Hofmann H., Interaction between the spike protein of human coronavirus NL63 and its cellular receptor ACE2. Adv. Exp. Med. Biol. 581, 281–284 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmann H., Marzi A., Gramberg T., Geier M., Pyrc K., van der Hoek L., Berkhout B., Pöhlmann S., Attachment factor and receptor engagement of SARS coronavirus and human coronavirus NL63. Adv. Exp. Med. Biol. 581, 219–227 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naskalska A., Dabrowska A., Nowak P., Szczepanski A., Jasik K., Milewska A., Ochman M., Zeglen S., Rajfur Z., Pyrc K., Novel coronavirus-like particles targeting cells lining the respiratory tract. PLOS ONE 13, e0203489 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naskalska A., Dabrowska A., Szczepanski A., Milewska A., Jasik K. P., Pyrc K., Membrane protein of human coronavirus NL63 is responsible for interaction with the adhesion receptor. J. Virol. 93, e00355-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milewska A., Zarebski M., Nowak P., Stozek K., Potempa J., Pyrc K., Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 88, 13221–13230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., Somasundaran M., Sullivan J. L., Luzuriaga K., Greenough T. C., Choe H., Farzan M., Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raj V. S., Mou H., Smits S. L., Dekkers D. H., Müller M. A., Dijkman R., Muth D., Demmers J. A., Zaki A., Fouchier R. A. M., Thiel V., Drosten C., Rottier P. J. M., Osterhaus A. D. M. E., Bosch B. J., Haagmans B. L., Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vlasak R., Luytjes W., Spaan W., Palese P., Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. U.S.A. 85, 4526–4529 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szczepanski A., Owczarek K., Bzowska M., Gula K., Drebot I., Ochman M., Maksym B., Rajfur Z., Mitchell J. A., Pyrc K., Canine respiratory coronavirus, bovine coronavirus, and human coronavirus OC43: Receptors and attachment factors. Viruses 11, 328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo P. C. Y., Lau S. K. P., Chu C.-m., Chan K.-h., Tsoi H.-w., Huang Y., Wong B. H. L., Poon R. W. S., Cai J. J., Luk W.-k., Poon L. L. M., Wong S. S. Y., Guan Y., Peiris J. S. M., Yuen K.-y., Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79, 884–895 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez S. R., Robinson C. C., Holmes K. V., Detection of four human coronaviruses in respiratory infections in children: A one-year study in Colorado. J. Med. Virol. 81, 1597–1604 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W., Wang W., Wang H., Lu R., Tan W., First infection by all four non-severe acute respiratory syndrome human coronaviruses takes place during childhood. BMC Infect. Dis. 13, 433 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dijkman R., Jebbink M. F., Koekkoek S. M., Deijs M., Jónsdóttir H. R., Molenkamp R., Ieven M., Goossens H., Thiel V., van der Hoek L., Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J. Virol. 87, 6081–6090 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominguez S. R., Shrivastava S., Berglund A., Qian Z., Góes L. G., Halpin R. A., Fedorova N., Ransier A., Weston P. A., Durigon E. L., Jerez J. A., Robinson C. C., Town C. D., Holmes K. V., Isolation, propagation, genome analysis and epidemiology of HKU1 betacoronaviruses. J. Gen. Virol. 95, 836–848 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominguez S. R., Travanty E. A., Qian Z., Mason R. J., Human coronavirus HKU1 infection of primary human type II alveolar epithelial cells: Cytopathic effects and innate immune response. PLOS ONE 8, e70129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pyrc K., Sims A. C., Dijkman R., Jebbink M., Long C., Deming D., Donaldson E., Vabret A., Baric R., van der Hoek L., Pickles R., Culturing the unculturable: Human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J. Virol. 84, 11255–11263 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X., Dong W., Milewska A., Golda A., Qi Y., Zhu Q. K., Marasco W. A., Baric R. S., Sims A. C., Pyrc K., Li W., Sui J., Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 89, 7202–7213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakkers M. J. G., Lang Y., Feitsma L. J., Hulswit R. J. G., de Poot S. A. H., van Vliet A. L. W., Margine I., de Groot-Mijnes J. D. F., van Kuppeveld F. J. M., Langereis M. A., Huizinga E. G., de Groot R. J., Betacoronavirus Adaptation to humans involved progressive loss of hemagglutinin-esterase lectin activity. Cell Host Microbe 21, 356–366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hulswit R. J. G., Lang Y., Bakkers M. J. G., Li W., Li Z., Schouten A., Ophorst B., van Kuppeveld F. J. M., Boons G.-J., Bosch B.-J., Huizinga E. G., de Groot R. J., Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. U.S.A. 116, 2681–2690 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallagher T. M., Escarmis C., Buchmeier M. J., Alteration of the pH dependence of coronavirus-induced cell fusion: Effect of mutations in the spike glycoprotein. J. Virol. 65, 1916–1928 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomura R., Kiyota A., Suzaki E., Kataoka K., Ohe Y., Miyamoto K., Senda T., Fujimoto T., Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J. Virol. 78, 8701–8708 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K., Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 81, 8722–8729 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Hamme E., Dewerchin H. L., Cornelissen E., Verhasselt B., Nauwynck H. J., Clathrin- and caveolae-independent entry of feline infectious peritonitis virus in monocytes depends on dynamin. J. Gen. Virol. 89, 2147–2156 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C., SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 18, 290–301 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milewska A., Nowak P., Owczarek K., Szczepanski A., Zarebski M., Hoang A., Berniak K., Wojarski J., Zeglen S., Baster Z., Rajfur Z., Pyrc K., Entry of human coronavirus NL63 into the cell. J. Virol. 92, e01933-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owczarek K., Szczepanski A., Milewska A., Baster Z., Rajfur Z., Sarna M., Pyrc K., Early events during human coronavirus OC43 entry to the cell. Sci. Rep. 8, 7124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J.-E., Cruz D. J. M., Shin H.-J., Receptor-bound porcine epidemic diarrhea virus spike protein cleaved by trypsin induces membrane fusion. Arch. Virol. 156, 1749–1756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Haan C. A. M., Haijema B. J., Schellen P., Wichgers Schreur P., te Lintelo E., Vennema H., Rottier P. J. M., Cleavage of group 1 coronavirus spike proteins: How furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J. Virol. 82, 6078–6083 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirchdoerfer R. N., Cottrell C. A., Wang N., Pallesen J., Yassine H. M., Turner H. L., Corbett K. S., Graham B. S., McLellan J. S., Ward A. B., Pre-fusion structure of a human coronavirus spike protein. Nature 531, 118–121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Haan C. A. M., Stadler K., Godeke G.-J., Bosch B. J., Rottier P. J. M., Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 78, 6048–6054 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Millet J. K., Whittaker G. R., Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 111, 15214–15219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada Y., Liu D. X., Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 83, 8744–8758 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kam Y.-W., Okumura Y., Kido H., Ng L. F. P., Bruzzone R., Altmeyer R., Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLOS ONE 4, e7870 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawase M., Shirato K., Matsuyama S., Taguchi F., Protease-mediated entry via the endosome of human coronavirus 229E. J. Virol. 83, 712–721 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu Z., Hingley S. T., Simmons G., Yu C., Das Sarma J., Bates P., Weiss S. R., Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 80, 5768–5776 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirato K., Kawase M., Matsuyama S., Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 87, 12552–12561 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simmons G., Gosalia D. N., Rennekamp A. J., Reeves J. D., Diamond S. L., Bates P., Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 102, 11876–11881 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., Welsch K., Winkler M., Schneider H., Hofmann-Winkler H., Thiel V., Pohlmann S., TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 87, 6150–6160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Krämer-Kühl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., von Hahn T., Simmons G., Hofmann H., Pöhlmann S., The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 87, 5502–5511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glowacka I., Bertram S., Müller M. A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T. S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S., Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85, 4122–4134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirato K., Matsuyama S., Ujike M., Taguchi F., Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J. Virol. 85, 7872–7880 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shirato K., Kawase M., Matsuyama S., Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology 517, 9–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harvey T. J., Hooper J. D., Myers S. A., Stephenson S.-A., Ashworth L. K., Clements J. A., Tissue-specific expression patterns and fine mapping of the human kallikrein (KLK) locus on proximal 19q13.4. J. Biol. Chem. 275, 37397–37406 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Shaw J. L. V., Diamandis E. P., Distribution of 15 human kallikreins in tissues and biological fluids. Clin. Chem. 53, 1423–1432 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Emami N., Diamandis E. P., New insights into the functional mechanisms and clinical applications of the kallikrein-related peptidase family. Mol. Oncol. 1, 269–287 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clements J., Hooper J., Dong Y., Harvey T., The expanded human kallikrein (KLK) gene family: Genomic organisation, tissue-specific expression and potential functions. Biol. Chem. 382, 5–14 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Kalinska M., Meyer-Hoffert U., Kantyka T., Potempa J., Kallikreins – the melting pot of activity and function. Biochimie 122, 270–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Memari N., Grass L., Nakamura T., Karakucuk I., Diamandis E. P., Human tissue kallikrein 9: Production of recombinant proteins and specific antibodies. Biol. Chem. 387, 733–740 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Memari N., Jiang W., Diamandis E. P., Luo L.-Y., Enzymatic properties of human kallikrein-related peptidase 12 (KLK12). Biol. Chem. 388, 427–435 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Darling M. R., Jackson-Boeters L., Daley T. D., Diamandis E. P., Human kallikrein 13 expression in salivary gland tumors. Int. J. Biol. Markers 21, 106–110 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Sotiropoulou G., Pampalakis G., Diamandis E. P., Functional roles of human kallikrein-related peptidases. J. Biol. Chem. 284, 32989–32994 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gruba N., Bielecka E., Wysocka M., Wojtysiak A., Brzezińska-Bodal M., Sychowska K., Kalińska M., Magoch M., Pęcak A., Falkowski K., Wiśniewska M., Sąsiadek L., Płaza K., Kroll E., Pejkovska A., Rehders M., Brix K., Dubin G., Kantyka T., Potempa J., Lesner A., Development of chemical tools to monitor human kallikrein 13 (KLK13) activity. Int. J. Mol. Sci. 20, 1557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyer-Hoffert U., Wu Z., Kantyka T., Fischer J., Latendorf T., Hansmann B., Bartels J., He Y., Gläser R., Schröder J.-M., Isolation of SPINK6 in human skin: Selective inhibitor of kallikrein-related peptidases. J. Biol. Chem. 285, 32174–32181 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kantyka T., Fischer J., Wu Z., Declercq W., Reiss K., Schröder J.-M., Meyer-Hoffert U., Inhibition of kallikrein-related peptidases by the serine protease inhibitor of Kazal-type 6. Peptides 32, 1187–1192 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Liu C., Ma Y., Yang Y., Zheng Y., Shang J., Zhou Y., Jiang S., Du L., Li J., Li F., Cell entry of porcine epidemic diarrhea coronavirus is activated by lysosomal proteases. J. Biol. Chem. 291, 24779–24786 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scorilas A., Borgoño C. A., Harbeck N., Dorn J., Schmalfeldt B., Schmitt M., Diamandis E. P., Human kallikrein 13 protein in ovarian cancer cytosols: A new favorable prognostic marker. J. Clin. Oncol. 22, 678–685 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Kapadia C., Chang A., Sotiropoulou G., Yousef G. M., Grass L., Soosaipillai A., Xing X., Howarth D. H., Diamandis E. P., Human kallikrein 13: Production and purification of recombinant protein and monoclonal and polyclonal antibodies, and development of a sensitive and specific immunofluorometric assay. Clin. Chem. 49, 77–86 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Ou X., Guan H., Qin B., Mu Z., Wojdyla J. A., Wang M., Dominguez S. R., Qian Z., Cui S., Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1. Nat. Commun. 8, 15216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ekholm I. E., Brattsand M., Egelrud T., Stratum corneum tryptic enzyme in normal epidermis: A missing link in the desquamation process? J. Invest. Dermatol. 114, 56–63 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Lundström A., Egelrud T., Stratum corneum chymotryptic enzyme: A proteinase which may be generally present in the stratum corneum and with a possible involvement in desquamation. Acta Derm. Venereol. 71, 471–474 (1991). [PubMed] [Google Scholar]
- 70.Bartlett J. D., Dental enamel development: Proteinases and their enamel matrix substrates. ISRN Dent 2013, 684607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cho A., Haruyama N., Hall B., Danton M. J. S., Zhang L., Arany P., Mooney D. J., Harichane Y., Goldberg M., Gibson C. W., Kulkarni A. B., TGF-ß regulates enamel mineralization and maturation through KLK4 expression. PLOS ONE 8, e82267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hart P. S., Hart T. C., Michalec M. D., Ryu O. H., Simmons D., Hong S., Wright J. T., Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J. Med. Genet. 41, 545–549 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Papagerakis P., Pannone G., Zheng L. I., Athanassiou-Papaefthymiou M., Yamakoshi Y., McGuff H. S., Shkeir O., Ghirtis K., Papagerakis S., Clinical significance of kallikrein-related peptidase-4 in oral cancer. Anticancer Res 35, 1861–1866 (2015). [PMC free article] [PubMed] [Google Scholar]
- 74.Charest-Morin X., Raghavan A., Charles M. L., Kolodka T., Bouthillier J., Jean M., Robbins M. S., Marceau F., Pharmacological effects of recombinant human tissue kallikrein on bradykinin B2 receptors. Pharmacol. Res. Perspect. 3, e00119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y., Luo W., Reiser G., Trypsin and trypsin-like proteases in the brain: Proteolysis and cellular functions. Cell. Mol. Life Sci. 65, 237–252 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ogawa K., Yamada T., Tsujioka Y., Taguchi J., Takahashi M., Tsuboi Y., Fujino Y., Nakajima M., Yamamoto T., Akatsu H., Mitsui S., Yamaguchi N., Localization of a novel type trypsin-like serine protease, neurosin, in brain tissues of Alzheimer’s disease and Parkinson’s disease. Psychiatry Clin. Neurosci. 54, 419–426 (2000). [DOI] [PubMed] [Google Scholar]
- 77.Lou J., Si H., Chen Y., Sun X., Zhang H., Niu A., Hu C., Correlation between KLK6 expression and the clinicopathological features of glioma. Contemp. Oncol. (Pozn) 18, 246–251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cerqueira C., Samperio Ventayol P., Vogeley C., Schelhaas M., Kallikrein-8 proteolytically processes human papillomaviruses in the extracellular space to facilitate entry into host cells. J. Virol. 89, 7038–7052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamilton B. S., Whittaker G. R., Cleavage activation of human-adapted influenza virus subtypes by kallikrein-related peptidases 5 and 12. J. Biol. Chem. 288, 17399–17407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Magnen M., Gueugnon F., Guillon A., Baranek T., Thibault V. C., Petit-Courty A., de Veer S. J., Harris J., Humbles A. A., Si-Tahar M., Courty Y., Kallikrein-related peptidase 5 contributes to H3N2 influenza virus infection in human lungs. J. Virol. 91, e00421-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garvin M. R., Alvarez C., Miller J. I., Prates E. T., Walker A. M., Amos B. K., Mast A. E., Justice A., Aronow B., Jacobson D., A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. eLife 9, e59177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van de Veerdonk F. L., Netea M. G., van Deuren M., van der Meer J. W. M., de Mast Q., Brüggemann R. J., van der Hoeven H., Kallikrein-kinin blockade in patients with COVID-19 to prevent acute respiratory distress syndrome. eLife 9, e57555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gibo J., Ito T., Kawabe K., Hisano T., Inoue M., Fujimori N., Oono T., Arita Y., Nawata H., Camostat mesilate attenuates pancreatic fibrosis via inhibition of monocytes and pancreatic stellate cells activity. Lab. Invest. 85, 75–89 (2005). [DOI] [PubMed] [Google Scholar]
- 84.Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R. Jr., Nunneley J. W., Barnard D., Pöhlmann S., McKerrow J. H., Renslo A. R., Simmons G., Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 116, 76–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matsuyama S., Shirato K., Kawase M., Terada Y., Kawachi K., Fukushi S., Kamitani W., Middle East respiratory syndrome coronavirus spike protein is not activated directly by cellular furin during viral entry into target cells. J. Virol. 92, e00683-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Souza L. R., Melo P. M., Paschoalin T., Carmona A. K., Kondo M., Hirata I. Y., Blaber M., Tersariol I., Takatsuka J., Juliano M. A., Juliano L., Gomes R. A., Puzer L., Human tissue kallikreins 3 and 5 can act as plasminogen activator releasing active plasmin. Biochem. Biophys. Res. Commun. 433, 333–337 (2013). [DOI] [PubMed] [Google Scholar]
- 87.Andrade D., Assis D. M., Santos J. A. N., Alves F. M., Hirata I. Y., Araujo M. S., Blaber S. I., Blaber M., Juliano M. A., Juliano L., Substrate specificity of kallikrein-related peptidase 13 activated by salts or glycosaminoglycans and a search for natural substrate candidates. Biochimie 93, 1701–1709 (2011). [DOI] [PubMed] [Google Scholar]
- 88.Lawrence M. G., Lai J., Clements J. A., Kallikreins on steroids: Structure, function, and hormonal regulation of prostate-specific antigen and the extended kallikrein locus. Endocr. Rev. 31, 407–446 (2010). [DOI] [PubMed] [Google Scholar]
- 89.Magklara A., Grass L., Diamandis E. P., Differential steroid hormone regulation of human glandular kallikrein (hK2) and prostate-specific antigen (PSA) in breast cancer cell lines. Breast Cancer Res. Treat. 59, 263–270 (2000). [DOI] [PubMed] [Google Scholar]
- 90.Morizane S., Yamasaki K., Kabigting F. D., Gallo R. L., Kallikrein expression and cathelicidin processing are independently controlled in keratinocytes by calcium, vitamin D3, and retinoic acid. J. Invest. Dermatol. 130, 1297–1306 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oikonomopoulou K., Hansen K. K., Saifeddine M., Vergnolle N., Tea I., Blaber M., Blaber S. I., Scarisbrick I., Diamandis E. P., Hollenberg M. D., Kallikrein-mediated cell signalling: Targeting proteinase-activated receptors (PARs). Biol. Chem. 387, 817–824 (2006). [DOI] [PubMed] [Google Scholar]
- 92.Hollenberg M. D., Oikonomopoulou K., Hansen K. K., Saifeddine M., Ramachandran R., Diamandis E. P., Kallikreins and proteinase-mediated signaling: Proteinase-activated receptors (PARs) and the pathophysiology of inflammatory diseases and cancer. Biol. Chem. 389, 643–651 (2008). [DOI] [PubMed] [Google Scholar]
- 93.Oikonomopoulou K., Hansen K. K., Saifeddine M., Tea I., Blaber M., Blaber S. I., Scarisbrick I., Andrade-Gordon P., Cottrell G. S., Bunnett N. W., Diamandis E. P., Hollenberg M. D., Proteinase-activated receptors, targets for kallikrein signaling. J. Biol. Chem. 281, 32095–32112 (2006). [DOI] [PubMed] [Google Scholar]
- 94.Seliga A., Lee M. H., Fernandes N. C., Zuluaga-Ramirez V., Didukh M., Persidsky Y., Potula R., Gallucci S., Sriram U., Kallikrein-kinin system suppresses type I interferon responses: A novel pathway of interferon regulation. Front. Immunol. 9, 156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Petraki C. D., Papanastasiou P. A., Karavana V. N., Diamandis E. P., Cellular distribution of human tissue kallikreins: Immunohistochemical localization. Biol. Chem. 387, 653–663 (2006). [DOI] [PubMed] [Google Scholar]
- 96.Bosch B. J., Bartelink W., P. J. M. Rottier, Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 82, 8887–8890 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Simmons G., Bertram S., Glowacka I., Steffen I., Chaipan C., Agudelo J., Lu K., Rennekamp A. J., Hofmann H., Bates P., Pöhlmann S., Different host cell proteases activate the SARS-coronavirus spike-protein for cell–cell and virus–cell fusion. Virology 413, 265–274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Heald-Sargent T., Gallagher T., Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses 4, 557–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Earnest J. T., Hantak M. P., Li K., McCray P. B. Jr., Perlman S., Gallagher T., The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLOS Pathog. 13, e1006546 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E., A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006). [DOI] [PubMed] [Google Scholar]
- 101.Horani A., Nath A., Wasserman M. G., Huang T., Brody S. L., Rho-associated protein kinase inhibition enhances airway epithelial Basal-cell proliferation and lentivirus transduction. Am. J. Respir. Cell Mol. Biol. 49, 341–347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Milewska A., Ciejka J., Kaminski K., Karewicz A., Bielska D., Zeglen S., Karolak W., Nowakowska M., Potempa J., Bosch B. J., Pyrc K., Szczubialka K., Novel polymeric inhibitors of HCoV-NL63. Antiviral Res. 97, 112–121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kovalski J. R., Bhaduri A., Zehnder A. M., Neela P. H., Che Y., Wozniak G. G., Khavari P. A., The functional proximal proteome of oncogenic ras includes mTORC2. Mol. Cell 73, 830–844.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lizama A. J., Andrade Y., Colivoro P., Sarmiento J., Matus C. E., Gonzalez C. B., Bhoola K. D., Ehrenfeld P., Figueroa C. D., Expression and bioregulation of the kallikrein-related peptidases family in the human neutrophil. Innate Immun. 21, 575–586 (2015). [DOI] [PubMed] [Google Scholar]
- 105.Milewska A., Kaminski K., Ciejka J., Kosowicz K., Zeglen S., Wojarski J., Nowakowska M., Szczubiałka K., Pyrc K., HTCC: Broad range inhibitor of coronavirus entry. PLOS ONE 11, e0156552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
stke.sciencemag.org/cgi/content/full/13/659/eaba9902/DC1
Fig. S1. Supplementation of RD cells with proteases does not result in a significant change in virus yield.
Table S1. KLK inhibitors used in the study.
Table S2. Primers used for the generation of plasmid constructs.
Table S3. Primers used for the PCR-based amplification of each KLK-encoding gene.
Table S4. Primers used in the CleavEx design.
File S1. Protein sequence data.