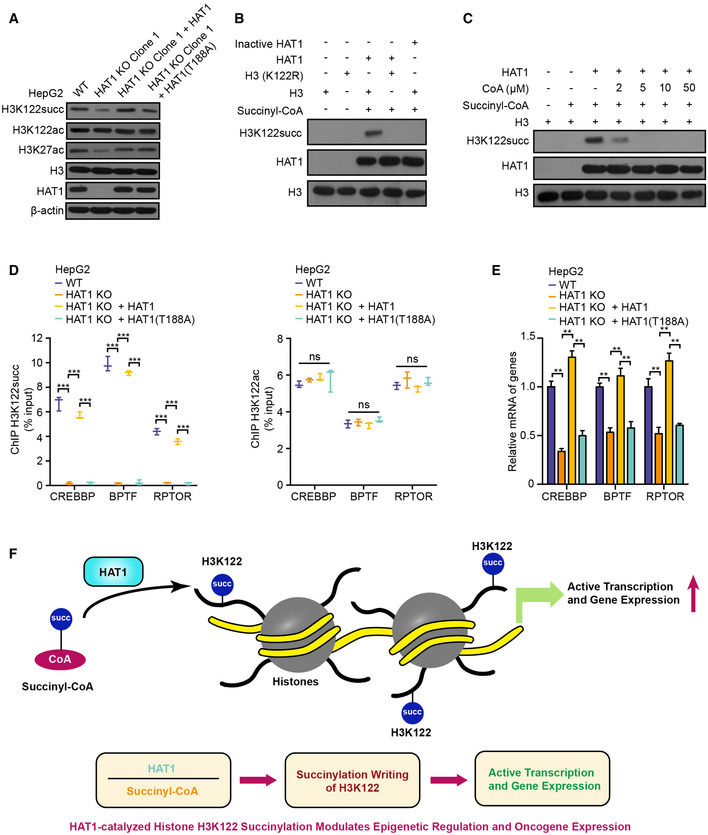
The levels of histone H3K122 succinylation, histone H3K122 acetylation, histone H3K27 acetylation, histone H3, HAT1, and β‐actin were examined by Western blot analysis in HepG2, HAT1 knockout (KO) Clone 1 HepG2 cells, and in HAT1 KO cells reconstituted with either HAT1 or mutant HAT1 (T188A).
HAT1‐catalyzed histone H3K122 succinylation was analyzed by mixing purified HAT1, histone H3 or histone H3 mutant (K122R), and succinyl‐CoA in the in vitro succinylation assays. Heat‐inactivated HAT1 was used as a negative control. Western blot analysis was performed with indicated antibodies.
HAT1‐catalyzed histone H3K122 succinylation was analyzed by mixing purified HAT1, histone H3, and succinyl‐CoA (2 μM) with or without the addition of the indicated concentrations of CoA. Western blot analysis was performed with indicated antibodies.
The effect of HAT1 on histone H3K122 succinylation and histone H3K122 acetylation at the indicated gene promoters was determined by ChIP assays with anti‐H3K122 succinylation antibody and anti‐H3K122 acetylation antibody and followed by quantitative PCR with primers of the promoter regions of CREBBP, BPTF, and RPTOR in WT HepG2 cells, HAT1 KO HepG2 cells, and HAT1 KO HepG2 cells reconstituted with either wild‐type HAT1 or mutant HAT1 (T188A). N = 3 biological replicates. Data are presented as mean ± SD. Student's t‐test, ***P < 0.001; ns, no significance.
The mRNA levels of CREBBP, BPTF, and RPTOR were measured by RT–qPCR in HepG2 cells, HAT1 KO HepG2 cells, and HAT1 KO HepG2 cells reconstituted with either wild‐type HAT1 or mutant HAT1 (T188A). N = 3 biological replicates. Data are presented as mean ± SD. Student's t‐test, **P < 0.01.
A model illustrating how HAT1‐catalyzed histone H3K122 succinylation modulates epigenetic regulation and gene expression.