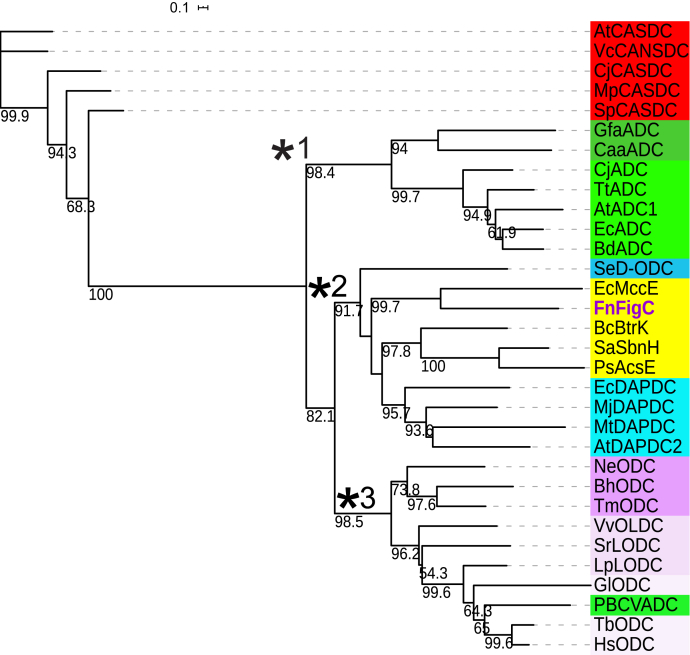
Figure 10.
Unrooted maximum likelihood tree of the alanine racemase-fold PLP-dependent decarboxylases. AtCASDC, Agrobacterium fabrum str. C58 (α-Proteobacteria) CASDC (NP_356481); VcCANSDC, Vibrio cholerae (γ-Proteobacteria) CANSDC (C6YCI2); CjCASDC, Campylobacter jejuni (ε-Proteobacteria) CASDC (WP_002877469); MpCASDC, Methanolacinia paynteri (Archaea, Euryarchaeota) CASDC (WP_048150085); GfaADC, Gramella forsetii (Bacteroidetes) ancestral short-form ADC (YP_863630); CaaADC, Chloroflexus aurantiacus (Chloroflexi) ancestral short-form ADC (YP_001634722); CjADC, Campylobacter jejuni (ε-Proteobacteria) long-form ADC (WP_002871443); TtADC, Thermus thermophilus (Deinococcus-Thermus) long-form ADC (AAS81619); AtADC1, Arabidopsis thaliana (Eukaryota, Viridiplantae) long-form ADC (AAB09723); EcADC, Escherichia coli (γ-Proteobacteria) long-form ADC (AAA24646); BdADC, Bacteroides dorei (Bacteroidetes) long-form ADC (AII67066); SeD-ODC, Salmonella enterica subsp. enterica serovar Typhimurium (γ-Proteobacteria) D-ODC (AAL21261); EcMccE, E. coli (γ-Proteobacteria) microcin C7 protein MccE (YP_006953769); FnFigC, Francisella tularensis subsp. novicida U112 (γ-Proteobacteria) citrylornithine decarboxylase FigC (ABF50970); BcBtrK, Bacillus circulans (Firmicutes) butirosin biosynthesis protein K (Q2L4H3); SaSbnH, Staphylococcus aureus (Firmicutes) citryl-L-2,3-diaminopropanoate decarboxylase (AAP82070); PsAcsE, Pseudomonas syringae (γ-Proteobacteria) O-citryl-L-serine decarboxylase (ELS42881); EcDAPDC, E. coli (γ-Proteobacteria) DAPDC (NP_417315); MjDAPDC, Methanocaldococcus jannaschii (archaea, euryarchaeota) DAPDC (Q58497); MtDAPDC, Mycobacterium tuberculosis (Actinobacteria) DAPDC (2O0T); AtDAPDC2, Arabidopsis thaliana (Eukaryota, Viridiplantae) DAPDC (Q94A94); NeODC, Nitrosomonas europaea (β-Proteobacteria) L-ODC (WP_011111520);BhODC, Bartonella henselae, (α-Proteobacteria) L-ODC (WP_011181090); TmODC, Thermotoga maritima (Thermotogae) L-ODC (NP_229669); VvOLDC, Vibrio vulnificus (γ-Proteobacteria) L-ornithine/L-lysine decarboxylase (AAO07938); SrLODC, Selenomonas ruminantium (Firmicutes) L-lysine/L-ornithine decarboxylase (BAA24923); LpLODC, Lupinus angustifolius (Eukaryota, Viridiplantae) L-lysine/L-ornithine decarboxylase (BAK32797); GlODC, Giardia lamblia (Eukaryota, Excavata) L-ornithine decarboxylase (EFO63849); PBCVADC, Paramecium bursaria chlorella virus 1 (chlorovirus) L-ODC-like ADC (NP_048554); TbODC, Trypanosoma brucei (Eukaryota, Excavata) L-ODC (AAA30218); HsODC, Homo sapiens (Eukaryota, Opisthokonta) L-ODC (AAA59967). The approximately 80 amino acid insertion in the long form ADCs relative to the other decarboxylases was removed, and all N termini and C termini sequences were trimmed to facilitate the alignment. ADC, arginine decarboxylase; CANSDC, L-carboxynorspermidine decarboxylase; CASDC, L-carboxyspermidine decarboxylase; DAPDC, meso-diaminopimelate decarboxylase; D-ODC, D-ornithine decarboxylase; L-ODC, L-ornithine decarboxylase.