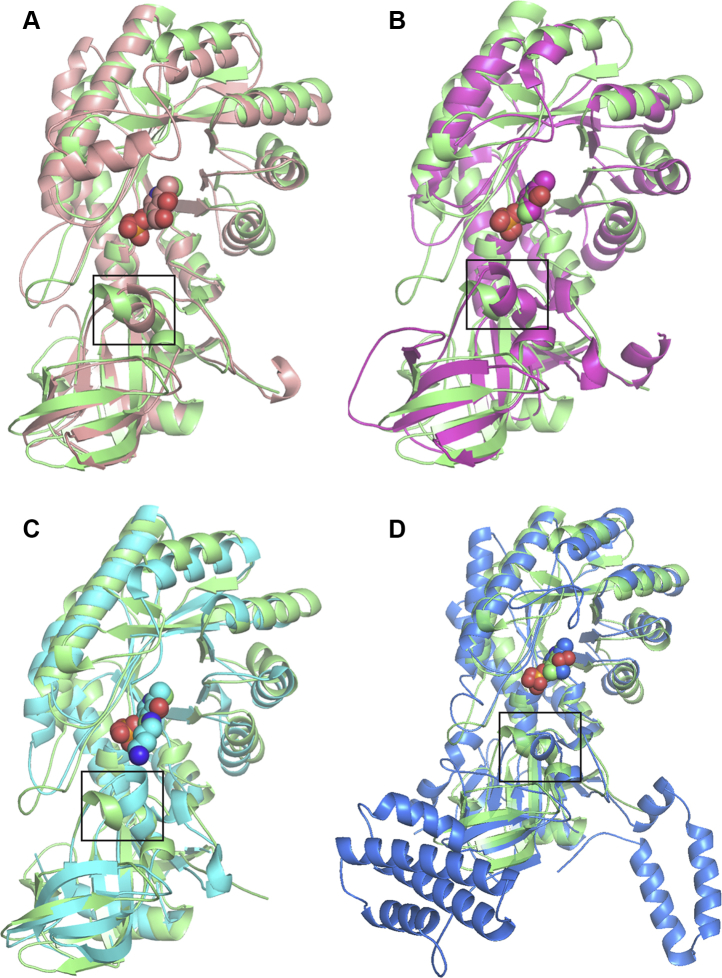
Figure 6.
Structural alignment of FigC with other PLP-dependent enzymes from the β/α structural fold.A, Methanocaldococcus jannaschii meso-diaminopimelate decarboxylase (1TWI) displayed in salmon. The RMS was 2.1 Å over 3866 atoms. B, Campylobacter jejuni carboxynorspermidine decarboxylase (3N29) displayed in magenta. The RMS was 2.2 Å over 3265 atoms. C, Vibrio vulnificus lysine/ornithine decarboxylase in complex with putrescine (2PLJ) displayed in turquoise for carbons and dark blue for the terminal amines. The RMS was 2.1 Å over 3147 atoms. D, V. vulnificus arginine decarboxylase (3N2O) displayed in dark blue. The RMS was 3.3 Å over 3050 atoms. Secondary structures are depicted as a cartoon and loops have been smoothed to simplify the display. FigC is shown in green in all structures, and the orientation is identical in all structures. The black box in each figure shows the specificity helix. PLP is shown as balls and is displayed for all structures, in addition the putrescine ligand is displayed for 2PLJ in panel C.