Abstract
Malstructured glycogen accumulates over time in Lafora disease (LD) and precipitates into Lafora bodies (LBs), leading to neurodegeneration and intractable fatal epilepsy. Constitutive reduction of glycogen synthase-1 (GYS1) activity prevents murine LD, but the effect of GYS1 reduction later in disease course is unknown. Our goal was to knock out Gys1 in laforin (Epm2a)-deficient LD mice after disease onset to determine whether LD can be halted in midcourse, or even reversed. We generated Epm2a-deficient LD mice with tamoxifen-inducible Cre-mediated Gys1 knockout. Tamoxifen was administered at 4 months and disease progression assessed at 12 months. We verified successful knockout at mRNA and protein levels using droplet digital PCR and Western blots. Glycogen determination and periodic acid–Schiff–diastase staining were used to analyze glycogen and LB accumulation. Immunohistochemistry using astrocytic (glial fibrillary acidic protein) and microglial (ionized calcium-binding adapter molecule 1) markers was performed to investigate neuroinflammation. In the disease-relevant organ, the brain, Gys1 mRNA levels were reduced by 85% and GYS1 protein depleted. Glycogen accumulation was halted at the 4-month level, while LB formation and neuroinflammation were significantly, though incompletely, prevented. Skeletal muscle analysis confirmed that Gys1 knockout inhibits glycogen and LB accumulation. However, tamoxifen-independent Cre recombination precluded determination of disease halting or reversal in this tissue. Our study shows that Gys1 knockdown is a powerful means to prevent LD progression, but this approach did not reduce brain glycogen or LBs to levels below those at the time of intervention. These data suggest that endogenous mechanisms to clear brain LBs are absent or, possibly, compromised in laforin-deficient murine LD.
Keywords: Lafora disease (Lafora progressive myoclonic epilepsy, MELF); glycogen; glycogen synthase; glycogen storage disease; neuroinflammation; neurodegenerative disease; Lafora bodies
Abbreviations: CAL, corpora amylacea–like; GFAP, glial fibrillary acidic protein; GYS1, glycogen synthase-1; IBA1, ionized calcium–binding adapter molecule 1; LB, Lafora body; LD, Lafora disease; PASD, periodic acid–Schiff–diastase; TAM, tamoxifen
Lafora disease (LD) is teenage-onset progressive myoclonus epilepsy that is typically fatal within 10 years of symptom onset (1, 2). LD is caused by autosomal recessive mutations in either EPM2A or NHLRC1 (also known as EPM2B), encoding laforin, a glucan phosphatase, and malin, an E3 ubiquitin ligase, respectively (3, 4, 5). The exact roles of both enzymes are unclear. It is, however, established that laforin and malin form a functional complex that regulates glycogen metabolism, with absence of either protein causing formation of malstructured glycogen (6, 7). LD is widely considered a glycogen storage disease (8, 9, 10, 11). The abnormal glycogen molecules precipitate and aggregate to form Lafora bodies (LBs), which continually accumulate, leading to symptom onset and then inexorably worsening neurodegeneration, seizures, and cognitive decline, culminating in vegetative state and death in status epilepticus and related complications (8).
Both laforin- and malin-deficient LD mouse models (12, 13, 14, 15) recapitulate the hallmarks of LD: glycogen accumulation, LB formation, and neuroinflammation (16, 17). In pursuit of a therapy for this intractable disease, one approach that emerged was to reduce glycogen synthase-1 (GYS1) activity, thereby interfering with chain elongation during glycogen synthesis, in order to prevent glycogen accumulation and LB formation. This approach proved successful with knockout of Gys1 or Ppp1r3c, encoding a protein (also known as PTG) involved in GYS1 activation. Both gene knockouts prevented LB formation, neurodegeneration, and other features of the disease in LD mice (16, 17, 18, 19). These results established proof of principle that targeting Gys1 for downregulation could be a promising therapeutic approach in LD. However, the mice in these studies had constitutively reduced GYS1. This left the question open as to how successful therapeutic targeting of Gys1 could be when LBs are already present, i.e., after disease onset and diagnosis.
We generated laforin-deficient (Epm2a−/−) mice with tamoxifen (TAM)-inducible Gys1 knockout. We administered TAM at 4 months, after murine disease onset, and assessed subsequent disease progression at 12 months. We analyzed glycogen content, LB accumulation, and neuroinflammation to determine whether LD progression can be halted in midcourse, or even potentially reversed.
Results
Laforin-deficient conditional Gys1 knockout mice
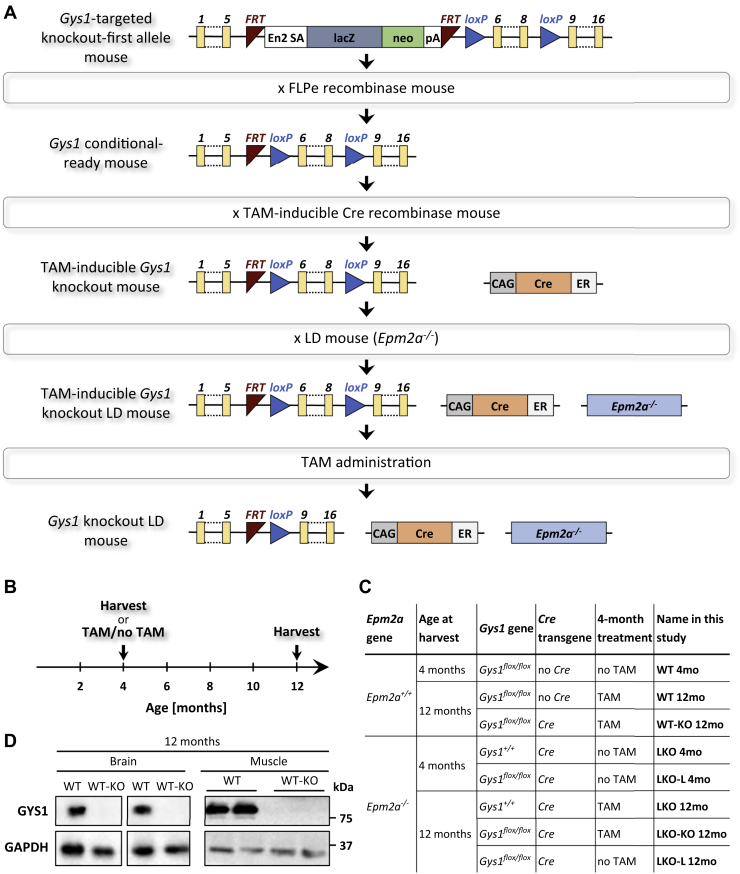
We generated conditional Gys1 knockout mice to test whether genetic pharmacotherapy targeting Gys1 following disease onset would lead to a halt or even reversal of LD progression. We used mice carrying a Gys1-targeting cassette with loxP sites flanking exons 6 to 8 together with TAM-inducible Cre to generate conditional Gys1-knockout mice in wild-type (WT) and Epm2a−/− backgrounds (Fig. 1A). A subset of mice was harvested at 4 months, while the majority of mice were either TAM-treated or not treated and then aged until 12 months (Fig. 1B). Figure 1C gives an overview of all experimental mice, not only explaining their genotype and treatments but also introducing the names of each group used throughout the study. In addition to the WT and Epm2a−/− control (WT, LKO) and Gys1 knockout (WT-KO, LKO-KO) mice, we included LKO-L mice at 4 and 12 months. These LKO-L mice carried floxed Gys1 and expressed Cre but were not TAM-treated. They served to evaluate whether TAM-independent Cre-mediated recombination (referred to as “Cre leakage”) occurred. At 12 months, WT-KO mice exhibited GYS1 protein knockdown in the brain and muscle (Fig. 1D), which clearly indicated that Cre-mediated Gys1 recombination and knockout was successful.
Figure 1.
Generation of Lafora disease (LD) model mice with tamoxifen (TAM)-inducible Gys1 knockout.A, overview of breeding strategy. B, schematic of study timeline. C, overview of experimental groups of mice. LKO, laforin (Epm2a) knockout; WT, wild-type. D, western blots depicting brain and muscle GYS1 protein levels with GAPDH as the loading control. Duplicates are biological replicates. CAG, chicken beta actin promoter/enhancer coupled with the cytomegalovirus immediate-early enhancer; En2 SA, mouse engrailed 2 splice acceptor sequence; ER, mutant estrogen receptor 1 ligand-binding domain; FRT, flippase recombination target; lacZ, reporter gene encoding β-galactosidase; loxP, Cre recombinase recognition site; neo, neomycin resistance gene; pA, SV40 polyadenylation signal.
Tamoxifen-induced Gys1 knockout halts glycogen and LB accumulation in the brain
To assess the effect of TAM treatment in the brain of conditional Gys1-knockout mice (WT-KO, LKO-KO) and potential Cre-leakage mice (LKO-L), we determined the frequency of recombination events by quantifying Gys1 mRNA that still contained exons 6 to 8. Levels of intact Gys1 mRNA were decreased by approximately 85% in both WT-KO and LKO-KO mice, while LKO-L mice showed no reduction and hence no Cre leakage at 4 or 12 months (Fig. 2A, Fig. S1A). Like WT-KO mice (Fig. 1D), LKO-KO mice exhibited strong knockdown of GYS1 protein levels, while no decrease in GYS1 was detected in LKO-L compared with LKO mice (Fig. 2B).
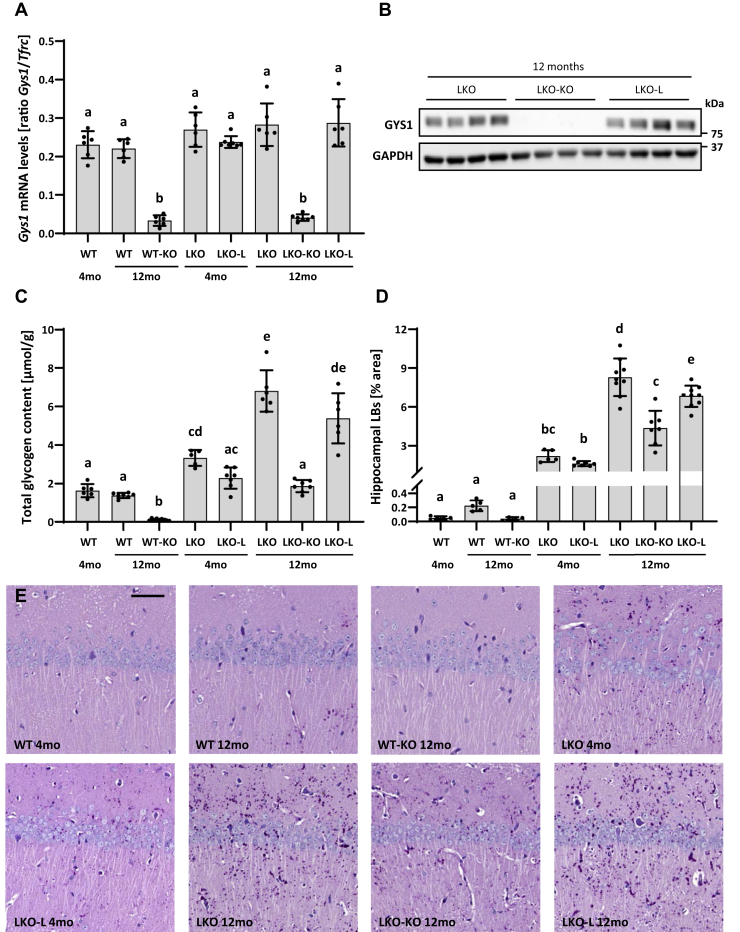
Figure 2.
Gys1 expression, glycogen, and Lafora body (LB) accumulation in the brain.A, Gys1 mRNA levels in the brain. Tfrc served as the reference gene. B, western blots depicting brain GYS1 protein levels with GAPDH as the loading control. Replicate lanes are from individual animals. C, total glycogen content in the brain. D, LB quantification in the hippocampus. Error bars indicate SD. Statistical significance (p < 0.05) is denoted by different letters, while lack of significance is reflected by at least one shared letter. Where applicable, subsets of experimental groups underwent secondary follow-up statistical testing (Fig. S1). E, representative images of the PASD-stained hippocampus. Scale bar: 50 μm. PASD, periodic acid–Schiff–diastase.
We then asked whether hallmarks of LD such as glycogen and LB accumulation in the brain were halted at the 4-month level, the time point of knockout induction, or perhaps even reversed. Glycogen determination confirmed an age-dependent increase in glycogen accumulation in LKO and LKO-L mice compared with WT in accordance with disease progression (Fig. 2C, Fig. S1B). There was a slight though not statistically significant trend toward less glycogen accumulation in LKO-L mice than in LKO control mice. Therefore, we directly compared LKO-KO mice with LKO-L mice in order to draw valid conclusions about disease halting or reversal. Doing so, we found clear stoppage in disease progression evidenced by LKO-L mice at 4 months having a very similar glycogen content as LKO-KO mice at 12 months (Fig. 2C, Fig. S1B). In WT-KO mice, glycogen levels were depleted, showing that Gys1 expression was sufficiently reduced to almost completely prevent synthesis of normal soluble glycogen. A similar reduction in soluble glycogen is to be expected in LKO-KO mice, and therefore, it can be concluded that the glycogen detected in these mice is mainly insoluble glycogen.
Hippocampal LB quantification showed a small though significant amount of corpora amylacea–like (CAL) bodies (normal age-related astrocytic glucan inclusions (20, 21)) in 12-month-old WT mice and age-dependent LB accumulation in LKO and LKO-L mice (Fig. 2D, Fig. S1C). CAL body formation was prevented by Gys1 knockout in WT-KO mice. LB accumulation was also significantly reduced in LKO-KO mice compared with LKO and LKO-L mice at 12 months, though slightly increased compared with LKO and LKO-L mice at 4 months. Therefore, Gys1 knockout at 4 months did not completely halt but significantly slowed LB formation in the hippocampus. In LD mice, the density of LBs is highest in the hippocampus, a structure whose pathologies commonly underlie epileptogenesis (22). However, in order to show a more complete picture regarding the LB load in the whole brain, we also quantified LBs in two additional large brain regions, cerebellum and cortex (Fig. S2, A–B). The results are very similar to what we saw in the hippocampus with the exception that in the cerebellum, a complete halt, versus only slowing of LB accumulation, could be achieved in LKO-KO mice. Representative periodic acid–Schiff–diastase (PASD) images are shown for LB visualization in Figure 2E and Fig. S2C.
Attenuated astrogliosis and microgliosis in the brain of conditional Gys1-knockout mice
Another hallmark of LD is glia-derived neuroinflammation (23). By performing immunohistochemistry for glial fibrillary acidic protein (GFAP) and ionized calcium–binding adapter molecule 1 (IBA1), we, respectively, assessed whether astrogliosis and/or microgliosis can be halted or reversed by targeting Gys1. In the hippocampus, WT and WT-KO mice did not exhibit any significant differences irrespective of Gys1 genotype or age (Fig. 3, Fig. S3). LKO and LKO-L mice showed minimal (GFAP) or no (IBA1) neuroinflammation compared with WT mice at 4 months but significantly increased GFAP and IBA1 signals by 12 months, confirming increased hippocampal neuroinflammation with LD progression. Twelve-month-old LKO-KO mice showed partial rescue with GFAP and IBA1 signal levels between 4-month-old and 12-month-old LKO and LKO-L mice levels.
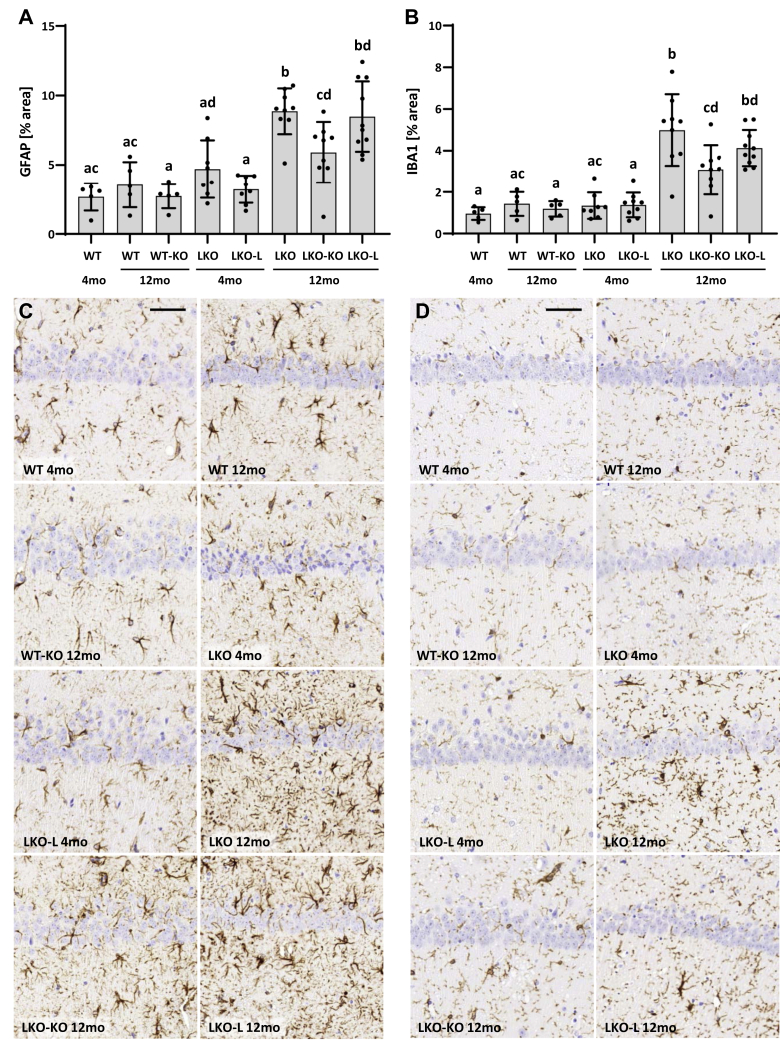
Figure 3.
Reactive gliosis in the brain.A–B, quantification of immunohistochemistry stain against GFAP (A) and IBA1 (B) in the hippocampus. Error bars indicate SD. Statistical significance (p < 0.05) is denoted by different letters, while lack of significance is reflected by at least one shared letter. Where applicable, subsets of experimental groups underwent secondary follow-up statistical testing (Fig. S3). C and D, representative images of GFAP (C) and IBA1 (D) immunohistochemistry in the hippocampus. Scale bars: 50 μm. GFAP, glial fibrillary acidic protein; IBA1, ionized calcium–binding adapter molecule 1.
GFAP and IBA1 signals were also quantified in the cerebellum (Fig. S4). However, astrogliosis was not significantly increased in LKO or LKO-L mice at 12 months, while microgliosis was significantly increased but not rescued in LKO-KO mice. In the cerebellum, LD-associated neuroinflammation was not as pronounced as in the hippocampus and consequently the timeframe to determine a halt in progression of neuroinflammation was too short to observe significant rescue in LKO-KO mice.
Prevention of the LD phenotype in the muscle
While LD manifests as a neurological disease, LBs additionally accumulate in murine and patient skeletal muscle. We therefore analyzed the muscle to further assess the effect of Gys1 knockout on glycogen and LB accumulation post disease onset. Like in the brain, we determined the extent of recombination events by quantifying the reduction of Gys1 mRNA with exons 6 to 8 still present (Fig. 4A, Fig. S5A). Surprisingly, Gys1 mRNA levels were already approximately 90% reduced in LKO-L mice at 4 months, indicating major Cre leakage in the muscle, which was not the case in the brain (Fig. 2A). At 12 months, Gys1 mRNA levels were further reduced by 3.5-fold in LKO-KO and by approximately 2-fold in LKO-L mice, culminating in 97% and 94% knockdown, respectively, while LKO control mice remained unaffected. GYS1 protein levels in LKO-KO mice were as reduced as in WT-KO mice at 12 months (Fig. 1E, Fig. 4B). However, we detected a comparable depletion of GYS1 levels in LKO-L mice at 12 months. Even at 4 months, GYS1 levels were already reduced in LKO-L mice, confirming Cre leakage at the protein level in addition to that seen at the mRNA level.
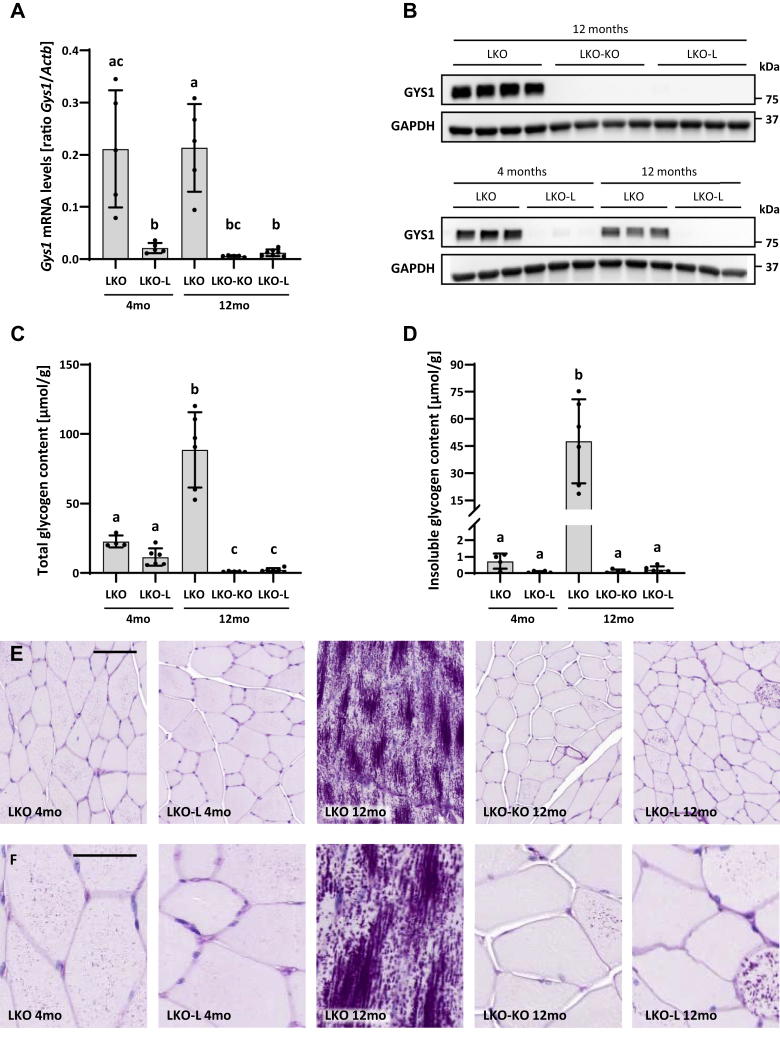
Figure 4.
Gys1 expression, glycogen, and Lafora body accumulation in the skeletal muscle.A, Gys1 mRNA levels in the muscle. Actb served as the reference gene. B, western blots depicting muscle GYS1 protein levels with GAPDH as the loading control. Replicate lanes are from individual animals. C, total glycogen content in the muscle. D, insoluble glycogen content in the muscle. Error bars indicate SD. Statistical significance (p < 0.05) is denoted by different letters, while lack of significance is reflected by at least one shared letter. Where applicable, subsets of experimental groups underwent secondary follow-up statistical testing (Fig. S5). E–F, representative images of the PASD-stained muscle. Scale bars: 100 μm (E); 50 μm (F). PASD, periodic acid–Schiff–diastase.
Further analyses focused on the effect of TAM-induced and TAM-independent Gys1 knockout on skeletal muscle total and insoluble glycogen content. For the latter, we used a recently established method that utilizes endogenously expressed metabolic enzymes to digest accessible soluble glycogen completely, leaving only the insoluble glycogen, which is not degradable (24). This method allowed us to determine whether changes in total glycogen content were mostly due to changes in soluble glycogen, insoluble glycogen, or both. LKO mice showed an increase in total and insoluble glycogen at 12 months, while the amount of insoluble glycogen was only approximately 3% of the total glycogen at 4 months (Fig. 4, C–D, Fig. S5, B–C). These results are in line with the extensive LB accumulation in LKO mice at 12 months as well as the comparatively small LB load at 4 months, with a substantial number of PASD-positive cells but very small bodies (Fig. 4, E–F). Already at 4 months, LKO-L mice exhibited 50% reduced levels of total glycogen and an almost complete depletion of insoluble glycogen, indicating that not only was disease progression attenuated but also levels of soluble glycogen were significantly reduced. Accordingly, fewer cells were PASD-positive with less/smaller LBs. The extent of LB formation in 12-month-old LKO-KO mice looked very similar to 4-month-old LKO-L mice, in accordance with the comparable insoluble glycogen content. However, in the LKO-KO mice, even the soluble glycogen was reduced further, as total glycogen was reduced by approximately 95% compared with 4-month-old LKO mice. We detected an approximately 2-fold higher amount of total and insoluble glycogen in LKO-L mice at 12 months than in LKO-KO mice. The slightly higher amount, though not statistically significant, is in line with the PASD results that show a few cells with relatively many LBs, which are probably cells where Cre leakage did not occur or only affected one of the two alleles. Taken together, our results show that Gys1 knockout prevented the LD muscle phenotype; however, significant Cre leakage occluded assessment of disease reversal in this organ.
Discussion
Hallmarks of LD are glycogen accumulation and formation of LBs, which are insoluble, glycogen-like particles, characterized by reduced branching and long chains (25). Therefore, reducing glycogen synthesis is an attractive avenue toward a therapy for the disease. From studying LD mouse models, we already know that reducing glycogen synthesis either by Gys1 knockout or indirectly by knockout of PTG prevents LB formation, neurodegeneration, and other features of the disease such as impaired autophagy (16, 17, 18, 19). Potential strategies to target brain glycogen synthesis are multiple, including antisense oligonucleotides, other forms of RNA interference, genome editing, and small-molecule inhibitors (8). Targeting glycogen synthesis as a therapy for LD has the advantage of applicability to patients with mutations in either EPM2A or NHLRC1, as opposed to gene replacement therapies, which would require separate development for each subgroup. A second major advantage is that downregulating a function is comparatively more practicable at the present time than replacing a missing function across the entire brain. Conventional knockout mouse models showed that it is possible to prevent LB formation and other disease-associated features when the capability to synthesize glycogen is blocked with the animal still in utero. Here, we asked the clinically more relevant question, namely, what would the effect of the intervention be after disease onset.
We generated a conditional Gys1-knockout mouse, using TAM-induced Cre-mediated recombination, which enabled us to downregulate glycogen synthesis in laforin-deficient mice at 4 months when the LD neuropathological phenotype is already clearly detectable. We studied the hallmarks of LD, glycogen accumulation, LB formation, and consequent neuroinflammation. Interestingly, total brain glycogen in LKO-KO at 12 months was significantly lower than in LKO control mice at 4 months. Although we hoped to see a reversal, our results rather show a halt in disease progression for the following two reasons. Firstly, we expect the amount of soluble glycogen in LKO-KO mice to be reduced as strongly as in WT-KO mice, where the overall glycogen content was reduced by approximately 90%, because Gys1 mRNA and protein levels suggest a similar extent of Cre recombination in WT-KO and LKO-KO mice. This indicates that we detected mostly insoluble glycogen in the Gys1-knockout mice (LKO-KO). Secondly, we need to take into consideration that the amount of total glycogen in 4-month-old LKO control mice is a combination of soluble and insoluble glycogen. In order to estimate the amount of insoluble (LB) glycogen at 4 months, we can subtract the amount of WT glycogen from the amount in LKO mice because we have shown that the amount of soluble glycogen is not altered in LD mice, at least in the muscle (26). This estimate gives approximately the same glycogen quantity as we see in LKO-KO mice at 12 months, further corroborating our conclusion of a halt in disease progression.
Strikingly, although still present, the halt in disease progression was not as complete when we look at the LB quantification results in the hippocampus. In the cerebellum, however, we observed a complete halt in LB formation which is more in line with our findings on total glycogen level, measured in whole brain samples. It could be that Cre recombination was slightly less efficient in the hippocampus than in other parts of the brain such as the cerebellum. The Gys1 mRNA and protein levels are an average over the whole brain and do not reveal any potential disparities in recombination efficiency in different brain regions. An alternative explanation for differences seen in different parts of the brain could be cell type–specific differences in Cre recombination, being, for instance, less efficient in astrocytes than in other cell types (e.g., neurons or other glial cells). It has been shown that a great number of LBs in LD mice are found in astrocytes (21, 27). Furthermore, it was found that the ratio between astrocytic and neuronal LBs can be different in different brain regions (21). Assuming this ratio is different in the hippocampus and cerebellum, cell type–specific differences in Cre recombination could explain the different extent of rescue seen in these two brain regions. Furthermore, glycogen metabolism in astrocytes is very different from glycogen metabolism in neurons (28, 29). These differences could result in different responses to reduced GYS1 levels. For example, GYS1 levels are much higher in astrocytes than in neurons, and it cannot be ruled out that the residual Gys1 expression in astrocytes, if, for example, only one allele is knocked out, could still drive LB formation at a slow pace, while Gys1 expression from only one allele in neurons may be too low to do so.
Neuroinflammation is a major feature of LD. As the disease progresses, reactive astrocytes and microglia appear and increase in number over time (16, 30, 31, 32), reflecting inflammatory and immune system responses. Intriguingly, 94% of the genes that are differentially expressed in both LD mouse models (Epm2a−/− and Nhlrc1−/−) are related to inflammatory and immune system responses, further validating the latter as critical to LD pathophysiology (23). Our GFAP and IBA1 immunohistochemistry results show slower progression of neuroinflammation in the hippocampus of LKO-KO mice, in accordance with the hippocampal LB quantification results. In the cerebellum, where a complete halt in LB formation was observed, less pronounced neuroinflammation in 12-month-old LKO and LKO-L mice rendered it hard to draw conclusions regarding rescue of neuroinflammation in this brain region. However, there is a strong association between epilepsy and hippocampal pathology (reviewed in (22)) and our data from hippocampus confirm that the severity of neuroinflammatory and immune responses is tightly linked to disease progression, as previously shown (23, 30).
The main focus in this study was the brain as the clinically relevant organ in LD. However, to further characterize our conditional Gys1 mouse model, we also analyzed skeletal muscle. Notably, we observed substantial TAM-independent Cre-mediated recombination in muscle, in contrast to the brain where Gys1 mRNA and GYS1 protein levels did not indicate significant Cre leakage. The original publication of the Cre mice used in this study reported Cre leakage in a small number of sporadic cells (33), and this commonly used mouse line has since accrued almost 600 references on Mouse Genome Informatics. Many of these publications did not report inclusion of appropriate controls to enable assessment of Cre leakage. Welle et al. (34) did assess Cre leakage and similarly reported a leakage-induced 50 to 90% reduction in target mRNA in skeletal muscle by 4 months of age.
TAM-inducible Cre mice, such as the ones used in this work, express transgenic Cre recombinase fused to a mutant estrogen receptor ligand-binding domain. Cre–estrogen receptor fusion proteins are sequestered in the cytoplasm until exposure to 4-hydroxytamoxifen (active TAM metabolite) enables translocation to the nucleus. Despite this putative mechanism for temporal regulation of Cre recombination, Cre leakage is a well-documented phenomenon caused by incomplete trapping of Cre in the cytoplasm (35, 36). Experimental parameters including Cre expression level and duration (37, 38, 39) and target gene sensitivity to recombination (40, 41) have also been associated with Cre leakage likelihood. Accordingly, the strong Cre promoter (chicken beta actin promoter/enhancer and cytomegalovirus immediate-early enhancer) in our mice and the 4-month pre-TAM timeframe may have contributed to Cre leakage. In regard to tissue-specific leakage, it should be noted that variability in both TAM-induced and TAM-independent recombination across tissues and cell types has been reported with the Cre mice used here, despite ubiquitous Cre expression (42, 43). Tissue-specific differences in the local chromatin structure at the locus of the Cre transgene (random integration), as well as that of the target gene, could result in increased Cre expression (44, 45) and a higher loxP site accessibility (36, 40), respectively, in the muscle. While both factors could promote TAM-independent Gys1 recombination, it cannot be ruled out that other unknown factors play a role. Overall, our muscle results provide further evidence that Cre leakage is a conserved and likely under-reported challenge of using TAM-inducible Cre regulation (45, 46). The difference in Cre leakage between the brain and muscle observed in this work highlights the importance of assessing Cre leakage in all tissues studied. Without including the LKO-L group, we would have overlooked Cre leakage in the muscle and our conclusions would have been very different—depicting reversal of LD in the muscle and conveying an incorrect clinically critical notion.
In the first months and even up to 3 years into the disease, patients with LD are neurologically still substantially preserved. Especially near onset, they are so healthy that they are nearly always misdiagnosed with the benign juvenile myoclonic epilepsy. As such, an approach that drastically slows or blocks disease progression, as we have shown in the present work in the Epm2a−/− mouse model of the disease, will be highly desirable. In the present era, patients with LD are diagnosed comparatively swiftly because EPM2A and NHLRC1 are included in most epilepsy gene panels, and therefore, most new patients would benefit. This study also aimed at uncovering whether downregulating glycogen synthesis could result in removal of existing LBs. Perhaps mechanisms such as autophagy no longer overwhelmed with constant LB formation could clear LBs accumulated to that point. Since we did not observe a net reduction in LBs despite GYS1 depletion, robust LB clearance mechanisms in the brain seem to be absent, at least in the laforin-deficient mouse model.
Several treatment approaches for LD have been suggested, and many are currently being probed in preclinical studies (8, 11). Some of these approaches aim at reducing GYS1 levels or activity. Those include administration of antisense oligonucleotides that promote degradation of Gys1 mRNA or of small-molecule inhibitors of GYS1. A recent publication describes the successful development of small-molecule inhibitors of GYS1 and demonstrates substantial inhibition of GYS1 in cell lysates (10). While in vivo efficacy remains to be proven, these strategies may eventually present viable treatment options for LD. However, if our results in mice translate to human LD, these GYS1-directed therapies may at most prevent further progression, but not reverse the disease. Therefore, GYS1-targeting therapies may have to be administered early in the disease progression with potentially limited benefits in patients with more advanced disease states. For the latter, other approaches in development, such as delivery of LB-digesting amylase to the brain (9), would be needed. There, the question will be to what degree the accompanying neuropathology will reverse following LB clearance.
Conclusions
Our results clearly show that slowing or halting LD progression after LBs have already formed is possible in the LD mouse model. Unfortunately, clearance of existing LBs was not observed. Our findings if translated to patients emphasize the importance of early diagnosis in treatments targeting glycogen synthesis downregulation.
Experimental procedures
Mice
Mice were housed with environmental enrichment in individually ventilated cages at 20 to 22 °C and had access to water and food ad libitum. Animals were sacrificed at 4 or 12 months of age by cervical dislocation. The brain and hind-limb muscle were harvested with half of each tissue immediately frozen in liquid nitrogen followed by −80 °C storage and the other half fixed in 10% buffered formalin followed by paraffin embedding. For brain analyses, both sexes were used with no indication of a disease phenotype–related sexual dimorphism, while only male mice were used for muscle analyses. Animal procedures were approved by The Centre for Phenogenomics Animal Care Committee and in compliance with the Canadian Council for Animal Care Guidelines.
Generation of laforin-deficient conditional Gys1-knockout mice
Mouse embryonic stem cells (C57BL/6N strain) containing a Gys1-targeted knockout-first allele with conditional potential (Gys1tm1a(EUCOMM)Wtsi; (47)) were obtained from the European Mouse Mutant Cell Repository. The Gys1tm1a(EUCOMM)Wtsi allele carries an FRT-flanked lacZ reporter and neomycin resistance sequence in intron 5 and loxP sites flanking exons 6 to 8 (Fig. 1A). Targeted embryonic stem cells were aggregated with morula and implanted in pseudo-pregnant females. Chimeric mice were crossed with B6N-Tyrc-Brd/BrdCrCrl mice to test for germline transmission. F1 mice were crossed with FLPe mice (B6;Cg-Tg(ACTFLPe)9205Dym/J; JAX stock #005703; (48)) to excise the FRT-flanked gene-trap cassette, yielding mice with a conditional-ready Gys1tm1c allele.
These Gys1tm1c mice were crossed with TAM-inducible Cre mice (B6.Cg-Tg(CAG-cre/Esr1∗)5Amc/J, JAX stock #004682; (33)). Upon TAM induction, Cre excises exons 6 to 8, leading to a premature termination codon after amino acid 293 (Gys1tm1d allele; full-length GYS1: 738 amino acids), which has been shown to be effective in knocking out Gys1 (49).
To assess therapeutic efficacy of Gys1 knockout after LD onset, mice with conditional-ready Gys1 and TAM-inducible Cre were crossed with laforin-deficient LD model mice (Epm2a−/−; (12)). See Table 1 for genotyping primers.
Table 1.
Genotyping primers
| Primer name | Purpose | Sequence [5’-3’] | Amplicon size [bp] |
|---|---|---|---|
| GeneTrap-F | Detection of gene trap cassette in knockout-first with conditional potential Gys1tm1a allele | AGGGTCAAGTAGCGGTGTTG | 207 |
| GeneTrap-R | GTGGGAAAGGGTTCGAAGTT | ||
| LoxP-F | Detection of loxP site in conditional-ready Gys1tm1c allele | AGGGTCAAGTAGCGGTGTTG | 270 (Gys1+), 458 (Gys1flox) |
| LoxP-R | TCAACTTCACAGGCAAAAACC | ||
| Cre-F | Detection of Cre transgene | GCGGTCTGGCAGTAAAAACTATC | 102 |
| Cre-R | GTGAAACAGCATTGCTGTCACTT | ||
| Epm2a-F | Detection of both Epm2a+ and Epm2a- alleles | GCATCGGCTGTAAGTTAGCC | 620 (Emp2a+), 430 (Epm2a-) |
| Epm2a-WT-R | Detection of Epm2a+ allele | CGTGTGTCCATTCTCCAGAA | |
| Epm2a-KO-R | Detection of Epm2a- allele | AGCGTATTCAATAACCCTTAAT |
Tamoxifen administration
TAM administration via intraperitoneal injection was determined as a reliable way to knock down Gys1. For intraperitoneal injections, 20 mg/ml of TAM-free base (Sigma, #T5648) was dissolved in corn oil (Sigma, #C8267) by rotating in a light-blocking tube overnight at 37 °C. Mice received three 0.225-mg/g injections evenly spaced across 5 to 8 days.
Gys1 expression analysis
RNA was extracted using the Qiagen RNeasy Lipid Tissue Mini Kit for the brain and RNeasy Mini Kit for the muscle (#74804, #74104). Extraction was performed according to the manufacturer’s instructions, using a 1-cc syringe and 21-g needle for tissue homogenization. DNase I digestion was performed to eliminate genomic DNA (DNase I Amplification Grade, Invitrogen, #18068-015 or DNase I, RNase-free, Thermo Scientific, #EN0521), followed by cDNA synthesis with SuperScript First-Strand Synthesis System (Invitrogen, #11904-01), using 1.45 μg of brain RNA and 0.73 μg of muscle RNA for this final step with a total volume of 20 μl.
Gys1 expression analysis was performed using the Bio-Rad QX200 Droplet Digital PCR (ddPCR) System. Transferrin receptor (Tfrc) was used as a reference gene in brain samples and actin beta (Actb) in muscle samples. For Gys1 and Tfrc, custom TaqMan assays were used (Gys1-F: CAGAGCAAAGCACGAATCCA, Gys1-probe: TTATGGGCACCTGGAC, Gys1-R: CATAGCGGCCAGCGATAAAG; Tfrc-F: TGCCTAATATACCTGTGCAAACAATC, Tfrc-probe: CAAGAGCTGCTGCAGAA, Tfrc-R: TTCCTTCCATTTTTCCAAATAGCT), while for Actb, a predesigned one (Mm01205647_g1, #4331182), all from Thermo Fisher Scientific. The Gys1 primer/probe set binds in exons 6 and 7 which are absent in Gys1-knockout mice. The 20-μl reaction mix consisted of 10 μl of 2× ddPCR SuperMix for Probes (Bio-Rad, #186-3023), 1 μl of Gys1 assay (FAM-labeled), 1 μl of reference gene assay (VIC-labeled), 0.5 μl (muscle) or 3 μl (brain) of cDNA, and 7.5 μl or 5 μl of nuclease-free water. Cycling conditions were 95 °C for 10 min, followed by 45 cycles of 94 °C for 30 s and 60 °C for 1 min, and eventually 98 °C for 10 min on a Life Technologies Veriti thermal cycler. Data were analyzed using QuantaSoft v1.4 (Bio-Rad). Gys1 was analyzed in duplex reaction mode together with the respective reference gene, and the results were expressed as the ratio of Gys1 to the reference gene. No-template controls were run in parallel with study samples.
Protein analysis
Lysate preparation and immunodetection of GYS1 and GAPDH was conducted as previously described (26).
Glycogen determination
Glycogen extraction and digestion using amyloglucosidase as well as glucose determination were performed as previously described (25). For analysis of insoluble muscle glycogen, ground tissue was incubated at 37 °C for 2.5 h prior to glycogen extraction in KOH (24).
Histological analysis
Paraffin-embedded brain or muscle tissue was sectioned and stained using PASD or immunohistochemical protocols at The Centre for Phenogenomics Pathology Services, Toronto. For immunohistochemistry, GFAP and IBA1 were used as neuroinflammation markers. Antibodies were diluted 1:2000 (rat anti-GFAP, Invitrogen, #130300) or 1:1500 (rabbit anti-IBA1, Wako, #01919741). Citrate buffer at pH 6 was used for antigen retrieval. Stained slides were scanned using a Pannoramic digital slide scanner (20× objective; 3DHistech), and representative images taken. LBs as well as GFAP and IBA1 signals were quantified in different parts of the brain using a method established in the image analysis program HistoQuant (3DHistech) by defining LBs or GFAP/IBA1 signals based on pixel color. Values are expressed as % area.
Statistics
Mice used in this study were assigned to one of eight experimental groups that differed in three dimensions: laforin genotype (WT or Epm2a−/−), age at harvest (4 or 12 months), and Gys1 genotype, which depended on the presence of Gys1 loxP sites, Cre transgene, and TAM administration (Fig. 1D). Quantitative data are represented as group averages with standard deviation (SD). All statistics were performed with GraphPad Prism 8.4.2. To allow for an appropriate statistical comparison of all eight groups, initial statistical analyses of dependent variables (e.g., brain Gys1 mRNA level, brain glycogen level) were performed by Welch’s one-way ANOVA (overall eight groups) followed by post hoc tests with Dunnett T3 multiple comparison correction. Statistically significant differences between means are assumed for p < 0.05. In main text figures, letters were used to denote if means were statistically different or not. Groups that have no letter in common are significantly different, while those that share at least one letter are statistically not different. In addition, where applicable, subsets of experimental groups underwent secondary follow-up testing. When analyzing effects of two specific dimensions at a time on dependent variables, two-way ANOVA was performed followed by post hoc tests with Bonferroni’s multiple comparison correction of p values. Figs. S1–S5 display means of groups compared in these secondary tests as well as significance levels as indicated. In cases where post hoc significance levels between two groups differ in the initial one-way ANOVA (overall eight groups) and the secondary ANOVAs (selected groups), statistical significance (letter code) in the main figures is in accordance with the secondary test.
Data availability
All data described are either presented in the article or are in the supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank C. Dalrymple for help with TAM administration, M. Ganguly for help with histological staining, and Dr T. Paton for help with ddPCR design.
Author contributions
S. N., E. E. C., F. N., and B. A. M. contributed to the conception and design of the study. S. N., E. E. C., and F. N. performed experiments and analyzed data. X. Z., S. G., S. P., D. R. G., A. M. P., P. W., and S. J. A. helped conduct experiments. S. N., F. N., and B. A. M. drafted and revised the manuscript. All authors reviewed and edited the manuscript.
Funding and additional information
This work was supported by families and friends of the Chelsea’s Hope Lafora Disease Research Fund, Associazione Italiana Lafora (AILA), France-Lafora, the Milana and Tatjana Gajic Lafora Disease Foundation, the Ontario Brain Institute, and the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (NIH) under award number P01 NS097197. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. B. A. M. holds the University of Texas Southwestern UTSW Jimmy Elizabeth Westcott Distinguished Chair in Pediatric Neurology.
Edited by Gerald Hart
Footnotes
This article contains supporting information.
Contributor Information
Felix Nitschke, Email: felix.nitschke@utsouthwestern.edu.
Berge A. Minassian, Email: berge.minassian@utsouthwestern.edu.
Supporting information
References
- 1.Delgado-Escueta A.V., Ganesh S., Yamakawa K. Advances in the genetics of progressive myoclonus epilepsy. Am. J. Med. Genet. 2001;106:129–138. doi: 10.1002/ajmg.1575. [DOI] [PubMed] [Google Scholar]
- 2.Minassian B.A. Lafora's disease: towards a clinical, pathologic, and molecular synthesis. Pediatr. Neurol. 2001;25:21–29. doi: 10.1016/s0887-8994(00)00276-9. [DOI] [PubMed] [Google Scholar]
- 3.Minassian B.A., Lee J.R., Herbrick J.A., Huizenga J., Soder S., Mungall A.J., Dunham I., Gardner R., Fong C.Y., Carpenter S., Jardim L., Satishchandra P., Andermann E., Snead O.C., 3rd, Lopes-Cendes I. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat. Genet. 1998;20:171–174. doi: 10.1038/2470. [DOI] [PubMed] [Google Scholar]
- 4.Serratosa J.M., Gomez-Garre P., Gallardo M.E., Anta B., de Bernabe D.B., Lindhout D., Augustijn P.B., Tassinari C.A., Malafosse R.M., Topcu M., Grid D., Dravet C., Berkovic S.F., de Cordoba S.R. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2) Hum. Mol. Genet. 1999;8:345–352. doi: 10.1093/hmg/8.2.345. [DOI] [PubMed] [Google Scholar]
- 5.Chan E.M., Young E.J., Ianzano L., Munteanu I., Zhao X., Christopoulos C.C., Avanzini G., Elia M., Ackerley C.A., Jovic N.J., Bohlega S., Andermann E., Rouleau G.A., Delgado-Escueta A.V., Minassian B.A. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat. Genet. 2003;35:125–127. doi: 10.1038/ng1238. [DOI] [PubMed] [Google Scholar]
- 6.Gentry M.S., Worby C.A., Dixon J.E. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8501–8506. doi: 10.1073/pnas.0503285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan M., Nitschke S., Steup M., Minassian B., Nitschke F. Pathogenesis of Lafora disease: transition of soluble glycogen to insoluble polyglucosan. Int. J. Mol. Sci. 2017;18:1743. doi: 10.3390/ijms18081743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nitschke F., Ahonen S.J., Nitschke S., Mitra S., Minassian B.A. Lafora disease - from pathogenesis to treatment strategies. Nat. Rev. Neurol. 2018;14:606–617. doi: 10.1038/s41582-018-0057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brewer M.K., Uittenbogaard A., Austin G.L., Segvich D.M., DePaoli-Roach A., Roach P.J., McCarthy J.J., Simmons Z.R., Brandon J.A., Zhou Z., Zeller J., Young L.E.A., Sun R.C., Pauly J.R., Aziz N.M. Targeting pathogenic Lafora bodies in Lafora disease using an antibody-enzyme fusion. Cell Metab. 2019;30:689–705.e686. doi: 10.1016/j.cmet.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang B., Frasinyuk M.S., Chikwana V.M., Mahalingan K.K., Morgan C.A., Segvich D.M., Bondarenko S.P., Mrug G.P., Wyrebek P., Watt D.S., DePaoli-Roach A.A., Roach P.J., Hurley T.D. Discovery and development of small-molecule inhibitors of glycogen synthase. J. Med. Chem. 2020;63:3538–3551. doi: 10.1021/acs.jmedchem.9b01851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentry M.S., Afawi Z., Armstrong D.D., Delgado-Escueta A., Goldberg Y.P., Grossman T.R., Guinovart J.J., Harris F., Hurley T.D., Michelucci R., Minassian B.A., Sanz P., Worby C.A., Serratosa J.M. The 5th International Lafora Epilepsy Workshop: basic science elucidating therapeutic options and preparing for therapies in the clinic. Epilepsy Behav. 2020;103:106839. doi: 10.1016/j.yebeh.2019.106839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganesh S., Delgado-Escueta A.V., Sakamoto T., Avila M.R., Machado-Salas J., Hoshii Y., Akagi T., Gomi H., Suzuki T., Amano K., Agarwala K.L., Hasegawa Y., Bai D.-S., Ishihara T., Hashikawa T. Targeted disruption of the Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum. Mol. Genet. 2002;11:1251–1262. doi: 10.1093/hmg/11.11.1251. [DOI] [PubMed] [Google Scholar]
- 13.DePaoli-Roach A.A., Tagliabracci V.S., Segvich D.M., Meyer C.M., Irimia J.M., Roach P.J. Genetic depletion of the malin E3 ubiquitin ligase in mice leads to Lafora bodies and the accumulation of insoluble laforin. J. Biol. Chem. 2010;285:25372–25381. doi: 10.1074/jbc.M110.148668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbull J., Wang P., Girard J.M., Ruggieri A., Wang T.J., Draginov A.G., Kameka A.P., Pencea N., Zhao X., Ackerley C.A., Minassian B.A. Glycogen hyperphosphorylation underlies lafora body formation. Ann. Neurol. 2010;68:925–933. doi: 10.1002/ana.22156. [DOI] [PubMed] [Google Scholar]
- 15.Criado O., Aguado C., Gayarre J., Duran-Trio L., Garcia-Cabrero A.M., Vernia S., San Millan B., Heredia M., Roma-Mateo C., Mouron S., Juana-Lopez L., Dominguez M., Navarro C., Serratosa J.M., Sanchez M. Lafora bodies and neurological defects in malin-deficient mice correlate with impaired autophagy. Hum. Mol. Genet. 2012;21:1521–1533. doi: 10.1093/hmg/ddr590. [DOI] [PubMed] [Google Scholar]
- 16.Turnbull J., DePaoli-Roach A.A., Zhao X., Cortez M.A., Pencea N., Tiberia E., Piliguian M., Roach P.J., Wang P., Ackerley C.A., Minassian B.A. PTG depletion removes Lafora bodies and rescues the fatal epilepsy of Lafora disease. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duran J., Gruart A., Garcia-Rocha M., Delgado-Garcia J.M., Guinovart J.J. Glycogen accumulation underlies neurodegeneration and autophagy impairment in Lafora disease. Hum. Mol. Genet. 2014;23:3147–3156. doi: 10.1093/hmg/ddu024. [DOI] [PubMed] [Google Scholar]
- 18.Pederson B.A., Turnbull J., Epp J.R., Weaver S.A., Zhao X., Pencea N., Roach P.J., Frankland P.W., Ackerley C.A., Minassian B.A. Inhibiting glycogen synthesis prevents Lafora disease in a mouse model. Ann. Neurol. 2013;74:297–300. doi: 10.1002/ana.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turnbull J., Epp J.R., Goldsmith D., Zhao X., Pencea N., Wang P., Frankland P.W., Ackerley C.A., Minassian B.A. PTG protein depletion rescues malin-deficient Lafora disease in mouse. Ann. Neurol. 2014;75:442–446. doi: 10.1002/ana.24104. [DOI] [PubMed] [Google Scholar]
- 20.Sinadinos C., Valles-Ortega J., Boulan L., Solsona E., Tevy M.F., Marquez M., Duran J., Lopez-Iglesias C., Calbo J., Blasco E., Pumarola M., Milan M., Guinovart J.J. Neuronal glycogen synthesis contributes to physiological aging. Aging Cell. 2014;13:935–945. doi: 10.1111/acel.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Augé E., Pelegrí C., Manich G., Cabezón I., Guinovart J.J., Duran J., Vilaplana J. Astrocytes and neurons produce distinct types of polyglucosan bodies in Lafora disease. Glia. 2018;66:2094–2107. doi: 10.1002/glia.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatzikonstantinou A. Epilepsy and the hippocampus. Front. Neurol. Neurosci. 2014;34:121–142. doi: 10.1159/000356435. [DOI] [PubMed] [Google Scholar]
- 23.Lahuerta M., Gonzalez D., Aguado C., Fathinajafabadi A., Garcia-Gimenez J.L., Moreno-Estelles M., Roma-Mateo C., Knecht E., Pallardo F.V., Sanz P. Reactive glia-derived neuroinflammation: a novel hallmark in Lafora progressive myoclonus epilepsy that progresses with age. Mol. Neurobiol. 2020;57:1607–1621. doi: 10.1007/s12035-019-01842-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nitschke S., Petkovic S., Ahonen S., Minassian B.A., Nitschke F. Sensitive quantification of α-glucans in mouse tissues, cell cultures, and human cerebrospinal fluid. J. Biol. Chem. 2020 doi: 10.1074/jbc.RA120.015061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitschke F., Sullivan M.A., Wang P., Zhao X., Chown E.E., Perri A.M., Israelian L., Juana-Lopez L., Bovolenta P., Rodriguez de Cordoba S., Steup M., Minassian B.A. Abnormal glycogen chain length pattern, not hyperphosphorylation, is critical in Lafora disease. EMBO Mol. Med. 2017;9:906–917. doi: 10.15252/emmm.201707608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan M.A., Nitschke S., Skwara E.P., Wang P., Zhao X., Pan X.S., Chown E.E., Wang T., Perri A.M., Lee J.P.Y., Vilaplana F., Minassian B.A., Nitschke F. Skeletal muscle glycogen chain length correlates with insolubility in mouse models of polyglucosan-associated neurodegenerative diseases. Cell Rep. 2019;27:1334–1344.e1336. doi: 10.1016/j.celrep.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubio-Villena C., Viana R., Bonet J., Garcia-Gimeno M.A., Casado M., Heredia M., Sanz P. Astrocytes: new players in progressive myoclonus epilepsy of Lafora type. Hum. Mol. Genet. 2018;27:1290–1300. doi: 10.1093/hmg/ddy044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saez I., Duran J., Sinadinos C., Beltran A., Yanes O., Tevy M.F., Martinez-Pons C., Milan M., Guinovart J.J. Neurons have an active glycogen metabolism that contributes to tolerance to hypoxia. J. Cereb. Blood Flow Metab. 2014;34:945–955. doi: 10.1038/jcbfm.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dienel G.A. The metabolic trinity, glucose-glycogen-lactate, links astrocytes and neurons in brain energetics, signaling, memory, and gene expression. Neurosci. Lett. 2015;637:18–25. doi: 10.1016/j.neulet.2015.02.052. [DOI] [PubMed] [Google Scholar]
- 30.López-González I., Viana R., Sanz P., Ferrer I. Inflammation in Lafora disease: evolution with disease progression in laforin and malin knock-out mouse models. Mol. Neurobiol. 2017;54:3119–3130. doi: 10.1007/s12035-016-9884-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rai A., Mishra R., Ganesh S. Suppression of leptin signaling reduces polyglucosan inclusions and seizure susceptibility in a mouse model for Lafora disease. Hum. Mol. Genet. 2017;26:4778–4785. doi: 10.1093/hmg/ddx357. [DOI] [PubMed] [Google Scholar]
- 32.Valles-Ortega J., Duran J., Garcia-Rocha M., Bosch C., Saez I., Pujadas L., Serafin A., Cañas X., Soriano E., Delgado-García J.M., Gruart A., Guinovart J.J. Neurodegeneration and functional impairments associated with glycogen synthase accumulation in a mouse model of Lafora disease. EMBO Mol. Med. 2011;3:667–681. doi: 10.1002/emmm.201100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi S., McMahon A.P. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 34.Welle S., Bhatt K., Pinkert C.A., Tawil R., Thornton C.A. Muscle growth after postdevelopmental myostatin gene knockout. Am. J. Physiol. Endocrinol. Metab. 2007;292:E985–E991. doi: 10.1152/ajpendo.00531.2006. [DOI] [PubMed] [Google Scholar]
- 35.Kristianto J., Johnson M.G., Zastrow R.K., Radcliff A.B., Blank R.D. Spontaneous recombinase activity of Cre-ERT2 in vivo. Transgenic Res. 2017;26:411–417. doi: 10.1007/s11248-017-0018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stifter S.A., Greter M. STOP floxing around: specificity and leakiness of inducible Cre/loxP systems. Eur. J. Immunol. 2020;50:338–341. doi: 10.1002/eji.202048546. [DOI] [PubMed] [Google Scholar]
- 37.Buelow B., Scharenberg A.M. Characterization of parameters required for effective use of tamoxifen-regulated recombination. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Hove H., Antunes A.R.P., De Vlaminck K., Scheyltjens I., Van Ginderachter J.A., Movahedi K. Identifying the variables that drive tamoxifen-independent CreERT2 recombination: implications for microglial fate mapping and gene deletions. Eur. J. Immunol. 2020;50:459–463. doi: 10.1002/eji.201948162. [DOI] [PubMed] [Google Scholar]
- 39.Chappell-Maor L., Kolesnikov M., Kim J.S., Shemer A., Haimon Z., Grozovski J., Boura-Halfon S., Masuda T., Prinz M., Jung S. Comparative analysis of CreER transgenic mice for the study of brain macrophages: a case study. Eur. J. Immunol. 2020;50:353–362. doi: 10.1002/eji.201948342. [DOI] [PubMed] [Google Scholar]
- 40.Vooijs M., Jonkers J., Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J., Willet S.G., Bankaitis E.D., Xu Y., Wright C.V.E., Gu G. Non-parallel recombination limits Cre-LoxP-based reporters as precise indicators of conditional genetic manipulation. Genesis. 2013;51:436–442. doi: 10.1002/dvg.22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donocoff R.S., Teteloshvili N., Chung H., Shoulson R., Creusot R.J. Optimization of tamoxifen-induced Cre activity and its effect on immune cell populations. Sci. Rep. 2020;10:15244. doi: 10.1038/s41598-020-72179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bult C., Blake J., Smith C., Kadin J., Richardson J., Mouse Genome Database Group . The Jackson Laboratory; Bar Harbor, ME: 2019. Mouse Genome Database (MGD) 2019 at the Mouse Genome Informatics Website. [Google Scholar]
- 44.Cotney J., Leng J., Oh S., DeMare L.E., Reilly S.K., Gerstein M.B., Noonan J.P. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 2012;22:1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long M.A., Rossi F.M.V. Silencing inhibits cre-mediated recombination of the Z/AP and Z/EG reporters in adult cells. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mingote S., Masson J., Gellman C., Thomsen G.M., Lin C.S., Merker R.J., Gaisler-Salomon I., Wang Y., Ernst R., Hen R., Rayport S. Genetic pharmacotherapy as an early CNS drug development strategy: testing glutaminase inhibition for schizophrenia treatment in adult mice. Front. Syst. Neurosci. 2016;9:165. doi: 10.3389/fnsys.2015.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T., Jackson D., Severin J., Biggs P., Fu J., Nefedov M. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 49.Duran J., Saez I., Gruart A., Guinovart J.J., Delgado-Garcia J.M. Impairment in long-term memory formation and learning-dependent synaptic plasticity in mice lacking glycogen synthase in the brain. J. Cereb. Blood Flow Metab. 2013;33:550–556. doi: 10.1038/jcbfm.2012.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described are either presented in the article or are in the supporting information.