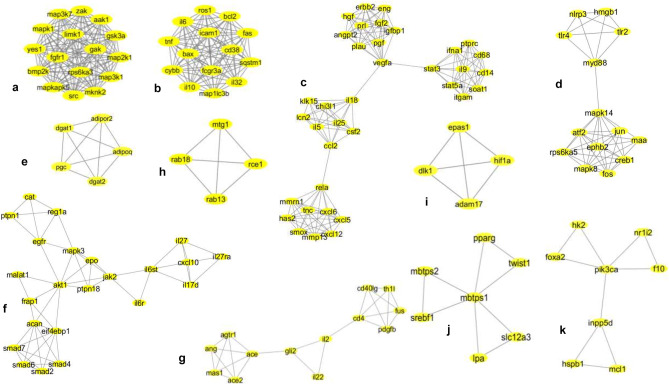
Fig. 3.
Network topologies of the most significant eleven MCODE clusters derived from association network of SARS-CoV-2 pathogenesis and probiotic treatment selected for further GO enrichment and pathway analysis. MCODE derived cluster 1: score 16, nodes 16 (src, limk1, rps6ka3, aak1, mapk1, mknk2, map3k1, gak, map2k1, fgfr1, mapkapk5, map3k7, bmp2k, zak, gsk3, yes1), and edges 120 (a). MCODE-derived cluster 2: score 14, nodes 14 (tnf, il6, map1lc3b, ros1, cd38, bax, fas, sqstm1, il10, fcgr3a, cybb, il32, bcl2, icam1), and edges 91 (b). MCODE-derived cluster 3: score 8.457, nodes 36 (cxcl5, cxcl12, tnc, has2, cxcl6, mmrn1, mmp13, smox, rela, ccl2, chi3l1, csf2, il5, il25, klk15, lcn2, il18, vegfa, eng, plau, pgf, prl, angpt2, igfbp1, hgf, erbb2, fgf2, stat3, stat5, ifna1, soat1, cd68, il9, itgam, ptprc, cd14), and edges 148 (c). MCODE-derived cluster 5: score 6.769, nodes 14 (tlr2, tlr4, nlrp3, myd88, hmgb1, mapk14, creb1, rps6ka5, jun, ephb2, mapk8, atf2, fos, maa), and edges 44 (d). MCODE-derived cluster 6: score 5, nodes 5 (dgat2, adipor2, dgat1, pgc, adipoq), and edges 10 (e). MCODE-derived cluster 9: score 4.818, nodes 23 (il27ra, il27, cxcl10, il17d, il6st, il6r, jak2, ptpn18, mapk3, epo, akt1, egfr, reg1a, ptpn1, cat, frap1, malat1, acan, eif4ebp1, smad4, smad6, smad2, smad7), and edges 53 (f). MCODE-derived cluster 10: score 4.167 and 4, nodes 13 (ace2, mas1, ace, ang, agtr1, il22, il2, gli2, cd4, cd40lg, fus, th1l, pdgfb) (g). MCODE-derived cluster 11: score 4, nodes 5 (rab18, rab13, mtg1, rce1), edges 25 (h). MCODE-derived cluster 13: score of 4, nodes 4 (hif1a, dlk1, adam17, epas1), and edges 6 (i). MCODE-derived cluster 35: score of 3, nodes 7 (pparg, twist1, lpa, slc12a, mbtps1, srebf1, mbtps2), and edges 9 (j). MCODE-derived cluster 38: score of 2.857, nodes 8 (hk2, nr1i2, foxa2, pik3ca, f10, inpp5d, hspb1, mcl1), and edges 10 (k)