Fig. 2. QTL mapping identifies a single large-effect locus explaining variation in matricidal hatching.
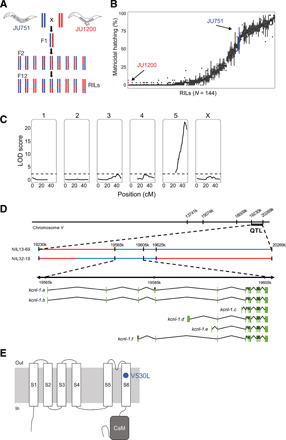
(A) Crossing scheme to generate F2 RILs derived from a parental cross between isolates JU751 and JU1200 (for details on phenotyping and genotyping of RILs, see fig. S3). (B) Phenotypic distribution (cumulative percentage of individuals undergoing matricidal hatching at mid-L4 + 72 hours at 15°C) of all replicates (n = 4 to 5) for each RIL (n = 144). Each boxplot displays all replicate values for a given RIL (x axis). Parental strains are colored in red (JU1200) and blue (JU751). (C) Linkage-mapping analysis of variation in matricidal hatching (proportion at mid-L4 + 72 hours). Each of the 146 SNP markers, separated by chromosome, was examined for correlation with phenotypic variation of the RIL panel (x-axis). The logarithm of the odds (LOD) score is shown on the y-axis. The dashed horizontal line shows the 5% significance threshold calculated with 1000 permutations. Phenotype data from 144 RILs were used for the mapping in R/qtl. (D) From QTL to QTN: Fine-mapping through NILs and additional genotyping of the target region identifies a single polymorphism in the gene kcnl-1, a small-conductance (SK) calcium-activated potassium channel subunit with six predicted isoforms, ranging from 2606 (isoform a) to 1937 (isoform c) amino acids. (E) Predicted structure of the KCNL-1 ion channel subunit (80). The KCNL-1 V530L variant in JU751 affects the predicted S6 transmembrane (TM) domain.
