Fig. 1. Acetaldehyde targets aa-tRNA and generates ethyl modification.
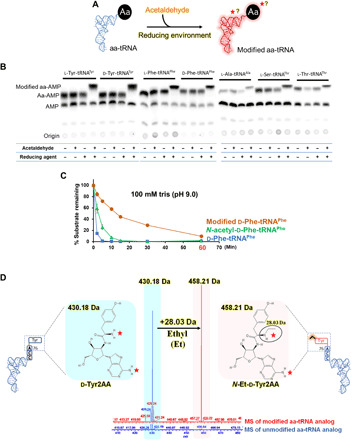
(A) Schematic showing reaction of aa-tRNA and acetaldehyde in the presence of a reducing agent. The red star or question mark indicates that the modification by acetaldehyde could be on amino acid or tRNA or both. (B) TLC is showing differences in the spot positions of aa-AMP corresponding to the unmodified and acetaldehyde-modified l-Tyr-tRNATyr, d-Tyr-tRNATyr, l-Phe-tRNAPhe, d-Phe-tRNAPhe, l-Ala-tRNAAla, l-Ser-tRNAThr, and l-Thr-tRNAThr [for each aa-tRNA, untreated aa-tRNA, aa-tRNA incubated with only acetaldehyde, aa-tRNA incubated with the only reducing agent, and aa-tRNA incubated with both acetaldehyde and reducing agent (in the manuscript, it is referred as “acetaldehyde-treated”) are shown in TLC]. (C) Alkali treatment of d-Phe-tRNAPhe, acetaldehyde-modified d-Phe-tRNAPhe, and N-acetyl-d-Phe-tRNAPhe is showing ultrastability of acetaldehyde-modified d-Phe-tRNAPhe. (D) ESI-MS analysis showing ethyl modification (a clear shift of 28.03 Da) on acetaldehyde-treated substrate analog of d-Tyr-tRNA [d-Tyr2AA (d-tyrosyl-2′-aminoadenosine)] [unmodified d-Tyr2AA with m/z = 430.18 (blue) and modified d-Tyr2AA with m/z = 458.21 (pink)].
