Fig. 3. NEDATs are readily acted upon by A. thaliana DTD2 but not by DTD1 or PTH.
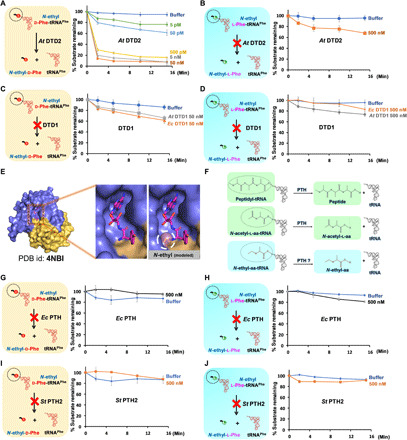
(A) Deacylation of N-ethyl-d-Phe-tRNAPhe (At) with At DTD2. (B) Deacylation of N-ethyl-l-Phe-tRNAPhe (At) with At DTD2. (C) Deacylation of N-ethyl-d-Phe-tRNAPhe with At DTD1 and Ec DTD1. (D) Deacylation of N-ethyl-l-Phe-tRNAPhe with At DTD1 and Ec DTD1. (E) Crystal structure of DTD1 in complex with d-Tyr3AA. Left of the zoomed-in section of the active site shows the binding mode of the snugly fit substrate; right shows a steric clash between the modeled ethyl modification on the α-NH2 group of d-amino acid and active site residues. PDB, Protein Data Bank. (F) Schematics showing subtle chemical differences among peptidyl-tRNA, N-acetyl-aa-tRNA, and NEAT (N-acetyl-aa-tRNAs differ with NEATs only by carbonyl oxygen present on the carbon that is attached to the α-NH2 group of amino acid of aa-tRNA). (G) Deacylation of N-ethyl-d-Phe-tRNAPhe (Ec) with Ec PTH. (H) Deacylation of N-ethyl-l-Phe-tRNAPhe (Ec) with Ec PTH. (I) Deacylation of N-ethyl-d-Phe-tRNAPhe (Pho) with St PTH2. (J) Deacylation of N-ethyl-l-Phe-tRNAPhe (Pho) with St PTH2. In all our biochemical assays, the concentration of substrate used was 0.2 μM. At, A. thaliana; Ec, E. coli; St, S. tokadii.
