Fig. 4. Acetaldehyde generates ethyl modification on d-aa-tRNA in vivo.
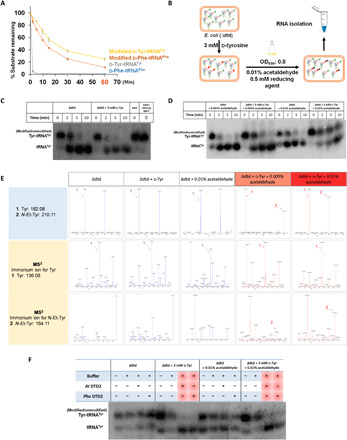
(A) CuSO4 + tris (8.0) treatment of in vitro generated d-Phe-tRNAPhe, d-Tyr-tRNATyr, modified (N-ethyl)-d-Phe-tRNAPhe, and modified (N-ethyl)-d-Tyr-tRNATyr. (B) Schematic showing the method used to generate NEDATs in E. coli. (C) Northern blot analysis showing no accumulation of Tyr-tRNATyr adducts in E. coli cells grown in the presence of only d-tyrosine. (D) Northern blot analysis showing the accumulation of Tyr-tRNATyr adducts in E. coli cells grown in the presence of d-tyrosine and acetaldehyde. (E) Alkali hydrolysis (incubation in 15% of NH4OH at 70°C for 18 hours) of aa-tRNA isolated from Δdtd E. coli, Δdtd E. coli + d-Tyr (grown in the presence of 3 mM d-tyrosine), and Δdtd E. coli + 0.01% acetaldehyde (grown in the presence of 0.01% acetaldehyde) yielded tyrosine peaks. N-ethyl-tyrosine peaks were seen only in Δdtd E. coli + 3 mM d-Tyr + 0.005% acetaldehyde (grown in the presence of 3 mM d-tyrosine and 0.005% acetaldehyde), and Δdtd E. coli + 3 mM d-Tyr + 0.01% acetaldehyde (grown in the presence of 3 mM d-tyrosine and 0.01% acetaldehyde). Fragmentation of the respective peaks also confirms the same. (F) Northern blot analysis showing deacylation of in vivo generated NEDATs by both archaeal DTD2 (Pho DTD2, 50 nM) and plant DTD2 (At DTD2, 50 nM).
