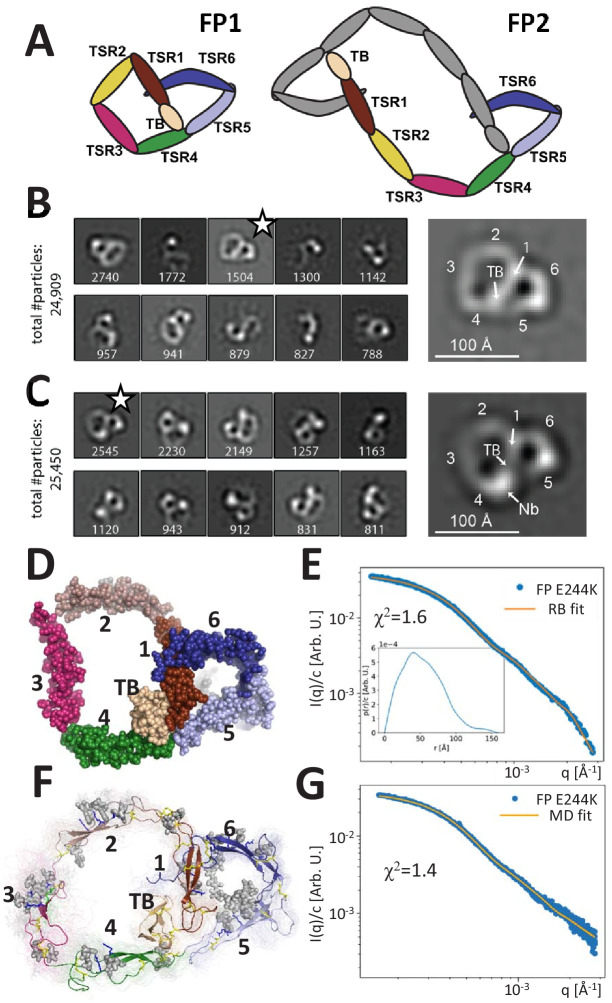
Figure 1. Principles of properdin architecture and the structure of FP1.
(A) Schematic representation of the FP1 monomer and FP2 as an example of an oligomer, where one subunit is colored gray, while the other is colored according to the domain structure as for FP1. (B) The 10 most populated nsEM 2D classes obtained with FP1 with the number of particles indicated. A magnified view of the 2D class marked by star is shown to the right. (C) As for (B), but for the FP1-hFPNb1 complex. Compared to FP1, an additional mass marks the location of hFPNb1 and hence TSR4 enabling assignment of the TB domain and the six thrombospondin repeats in the magnified view to the right. (D) Representative atomic model of FP1 E244K derived by rigid-body modeling against the SAXS data. (E) Comparison of SAXS experimental data (Pedersen et al., 2017) and the fitted scattering curve corresponding to the FP1 E244K model presented in (E). Insert to (E): p(r) function derived from the SAXS data. (F) Conformational ensemble of FP1 E244K sampled by a 1 μs MD simulation represented by 100 frames with 10 ns interval shown as transparent tubes. The starting model is displayed as a cartoon with the glycans and glycosylated residues in gray stick representation. Disulfide bridges are represented by yellow sticks. (G) Comparison of SAXS experimental data and the scattering curve obtained from MD ensemble after refinement using the Bayesian maximum entropy approach, the two curves fit with χ2 = 1.4. The minor difference in the experimental data apparent at the highest q-values in (E) and (G) is due to subtraction of a constant by CORAL in (E).