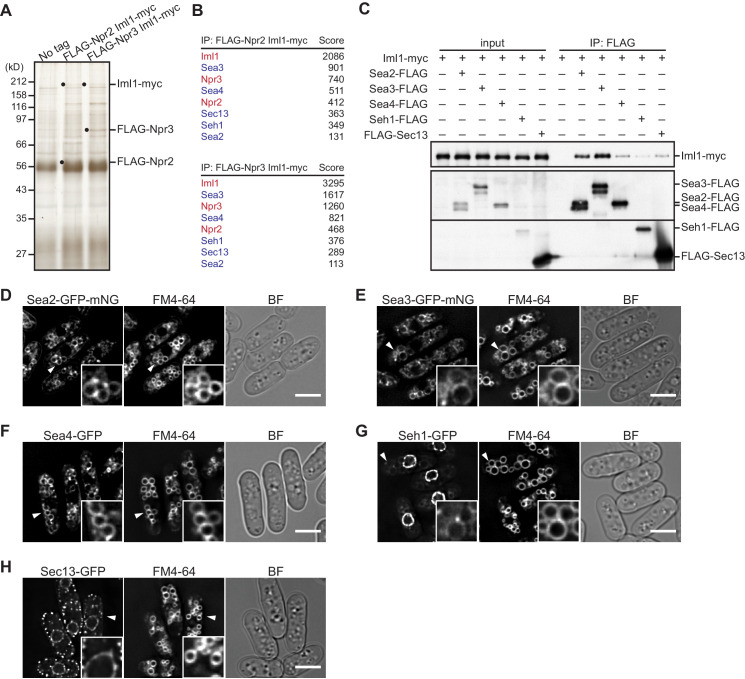
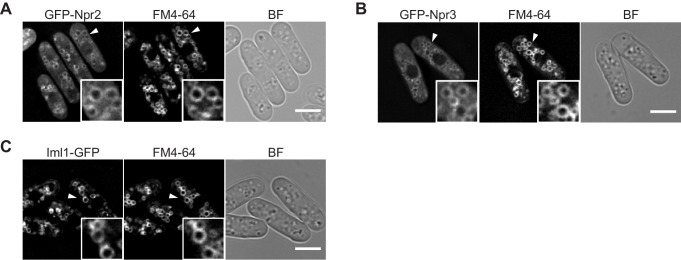
Figure 1. Identification of GATOR2 components as GATOR1-binding proteins.
(A) Affinity purification of the GATOR complex in fission yeast. The GATOR1 complex was purified from cells co-expressing FLAG-Npr2 and Iml1-myc, as well as those co-expressing FLAG-Npr3 and Iml1-myc, by two successive immunoprecipitation steps using anti-FLAG and anti-myc beads. The immunoprecipitates were resolved on SDS–PAGE followed by silver staining. The protein bands corresponding to the tagged Iml1, Npr2, and Npr3 are indicated by black dots. Wild-type cells expressing untagged proteins were used as a control (No tag). (B) Proteins co-purified with the GATOR1 subunits in (A) were subjected to mass spectrometric analysis. For each protein listed in the tables, two or more peptides were identified. The sum of peptide scores that exceed the 95% confidence level (p<0.05) is also shown for each of the identified proteins. GATOR1 and GATOR2 components are indicated in red and blue, respectively. (C) Physical interactions between Iml1 and the GATOR2 components were confirmed. Crude lysates (input) were prepared from the iml1:myc strain expressing one of the GATOR2 components tagged with FLAG for immunoprecipitation. The anti-FLAG immunoprecipitates (IP: FLAG) were analyzed by immunoblotting. (D–H) The cellular localization of GATOR2 proteins. The indicated fluorescence-tagged strains were grown in Edinburgh minimal medium (EMM) at 30°C for microscopy. Vacuolar membranes were visualized by the fluorescent dye FM4-64. Z-axial images were collected, and mid-section images after deconvolution are shown. Insets show the magnified views of the marked areas. BF, bright-field image. Bars, 5 μm.