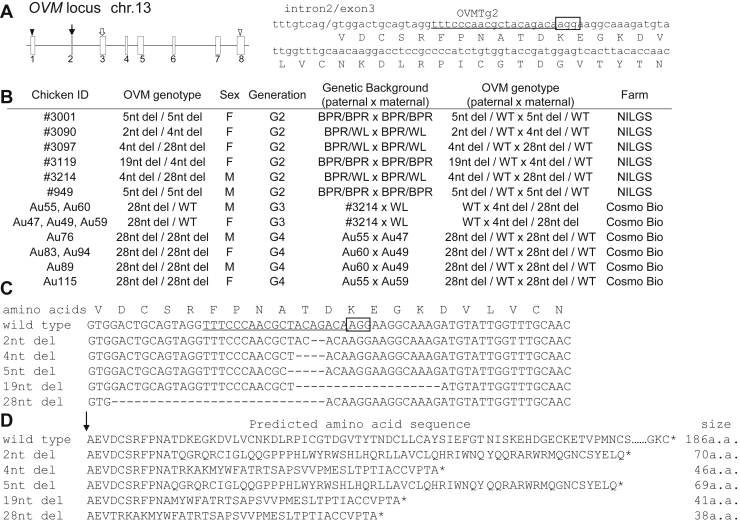
Figure 1.
OVM mutation in gene-targeted chickens. (A) Schematic representation of sgRNA targeting OVM. Left panel, exon-intron organization of OVM. Boxes with numbers indicate OVM exons. Closed and open triangles represent positions of start and stop codons, respectively. A closed arrow indicates the location of the peptide cleavage site region. The open arrow indicates the targeted OVM site. Right panel, OVM target sequence in exon 3. Lowercase and uppercase letters refer to DNA and amino acid sequences, respectively. Underlined and boxed text refer to sgRNA targeting site and adjoining putative protospacer adjacent motif (PAM) sequence, respectively. The slash indicates the boundary of intron 2 and exon 3. (B) OVM genotype of each biallelic mutant chicken in this study. F, female; M, male; WT, wild-type; nt del, nucleotide deletions; BPR, Barred Plymouth Rock; WL, White Leghorn. (C) Sequence analysis of CRISPR/Cas9-mediated deletion mutations in gene-targeted chickens. Wild-type DNA and amino acid sequences are shown in the top rows. The guide RNA targeting site and PAM sequence are underlined and boxed, respectively. The number of deleted nucleotides (2–28) is indicated to the left of each sequence. Deleted nucleotides are shown as dashes. (D) Wild-type amino acid sequences, and amino acid sequences predicted from each mutant DNA sequence. Predicted amino acid size is shown to the right of each sequence. Arrow shows the cleavage site in OVM.