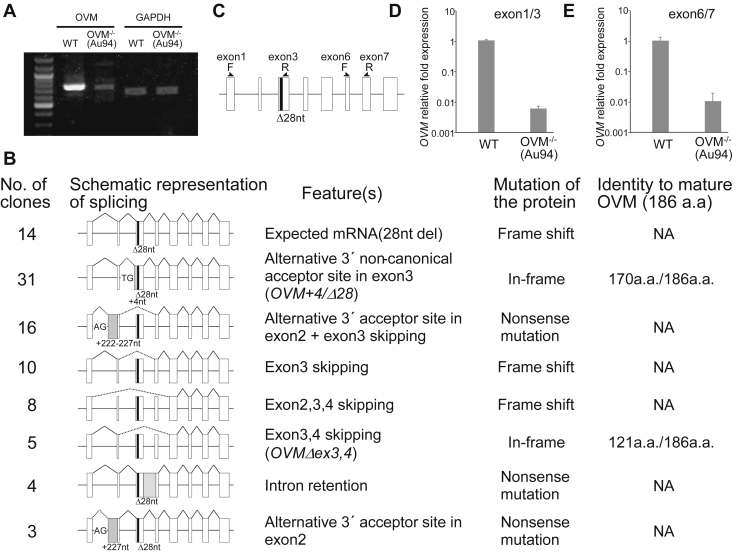
Figure 2.
Analysis of OVM transcripts expressed in the oviduct of OVM−/− hens. (A) RT-PCR analysis of OVM mRNA in WT and OVM−/− oviduct samples. Primers in exons 1 and 8 of OVM were used (see Materials and Methods). Internal control was GAPDH. (B) Summary of all OVM mRNA variants identified in OVM−/− hens. Schematic represents different types of pre-RNA splicing events. Open boxes represent OVM exons. Solid and dashed lines indicate normal and abnormal splicing, respectively. The black box represents the 28-nucleotide deletion in exon 3; gray boxes indicate aberrant exons. Alternative 3′ acceptor sites (noncanonical TG and canonical AG) are shown. NA, not applicable. (C) Schematic representation of primer positions for quantitative RT-PCR. Two primer sets were designed to quantify OVM transcripts. (D, E) Expression levels of OVM transcripts detected with quantitative RT-PCR. (D) Primers corresponding to exons 1 and 3, and (E) exons 6 and 7 were used. Error bars indicate standard deviation.