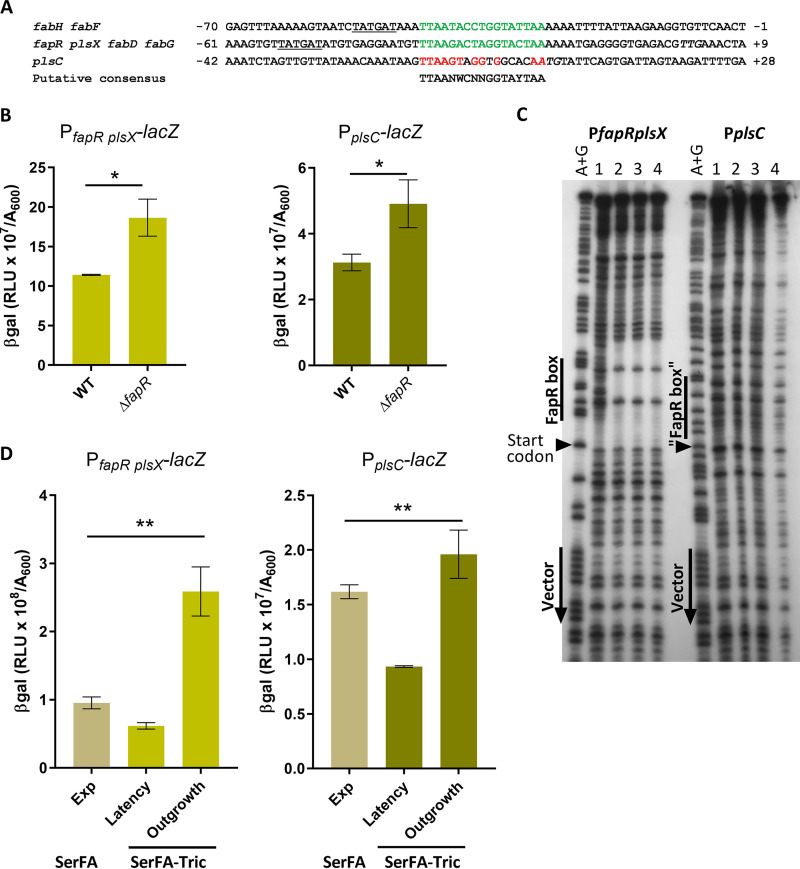
FIG 5.
FapR binds the plsX but not the plsC promoter region, but affects expression of both genes. (A) Sequence alignment of FapR-binding sites in S. aureus as published (13). Positions correspond to the predicted start codon of the first open reading frame (ORF) in each operon. Confirmed 17-bp FapR-binding sequences are in green. The presumed conserved FapR-binding nucleotides upstream of the plsC start site are in red. In the consensus sequence below the alignment, N indicates any nucleotide; W indicates A or T; Y indicates T or C. The putative −10 RNA polymerase binding sites are underlined (13). The plsC ATG and fapR TTG start sites are in italics. (B) Expression of PfapR plsX-lacZ and PplsC-lacZ fusions is upregulated in the ΔfapR strain. Strains were grown to A600 = ∼1.5 in SerFA medium prior to β-gal determinations. Data presented are means ± standard deviations from three biological replicates; *, P ≤ 0.05, ns, not significant, using Mann-Whitney. (C) DNase I footprinting assay of the PfapRplsX and PplsC promoter regions with FapR. Radiolabeled PCR fragments corresponding to PplsX and PplsC were used as DNA targets. Various amounts of FapR (lane 1, 0 nmol; lane 2, 1.2 nmol; lane 3, 6 nmol; and lane 4, 12 nmol) were incubated with 0.2 pmol DNA before DNase 1 digestion. Lane A+G contains the Maxam and Gilbert reaction of the labeled strand. Positions of vector and "FapR box" sequences are indicated by vertical lines, plsX and plsC start codons are indicated by black arrows. (D) Expression of PfapR plsX-lacZ and PplsC-lacZ fusions in S. aureus Newman strain during growth in nonselective (SerFA) and anti-FASII (SerFA-Tric) media. β-Gal values are shown for samples processed in SerFA at optical density at 600 nm (OD600) = ∼2.0 (exponential phase [Exp]) and in SerFA-Tric after 3 h in latency (OD600 = ∼0.3) and upon adaptation outgrowth at 17 h (OD600 = ∼2.0 [Outgrowth]). Means and standard deviations are shown for three independent experiments. P values were determined by Kruskal-Wallis test; **, P ≤ 0.005; ns, not significant.