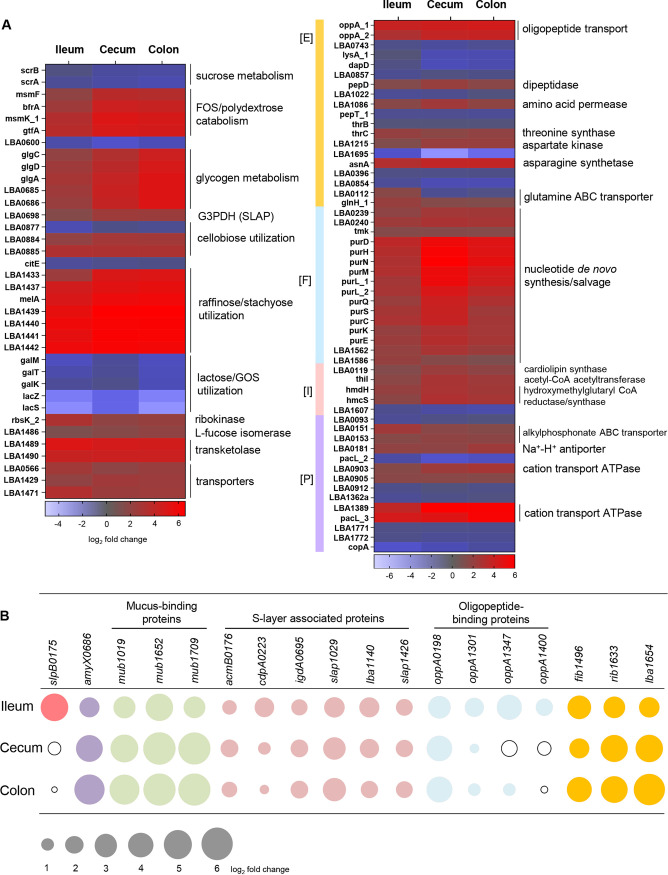
FIG 3.
(A) In vivo, differentially regulated core genes in L. acidophilus NCFM involved in carbohydrate [G] (left panel), amino acid [E], nucleotide [F], lipid [I], and inorganic ion transport and metabolism [P] (right panel) (see Table S5 at Dryad, https://doi.org/10.5061/dryad.mcvdncjzr). (B) Ileum-induced genes of cell surface proteins and their respective expression levels in the cecum and colon. Fold change of induction corresponds to bubble size, and downregulated fold change is represented by unfilled bubbles. Log2 fold change, differential expression ratio in ileum, cecum, or colon versus stationary-phase cells resuspended in PBS prior to oral gavage (baseline transcriptome on day 0, in vitro). G3PDH, glyceraldehyde-3-phosphate dehydrogenase; CoA, coenzyme A.