Figure 3. Cpa thioester bond cleavage is negatively force-dependent.
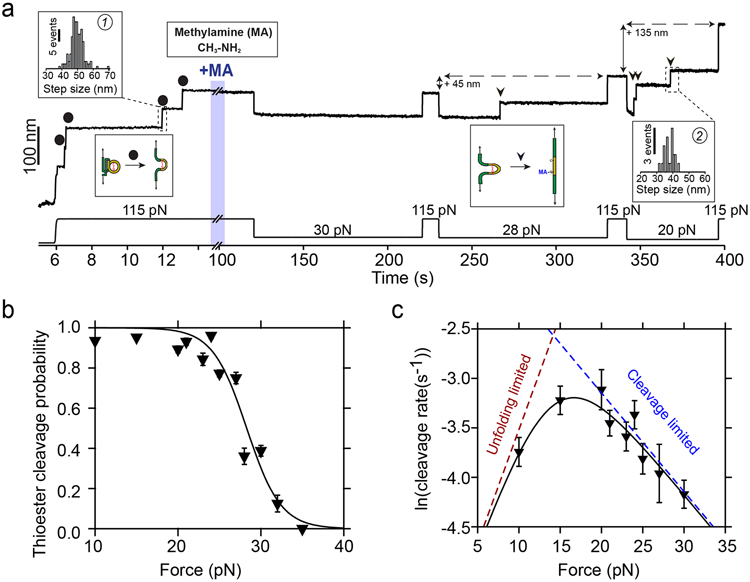
a) Magnetic tweezers trajectory of the Cpa polyprotein. After the unfolding of the thioester-intact Cpa domains at 115 pN (circles; histogram inset #1: 49.6±4.1 nm, mean±SD, n=164), the buffer is exchanged to a Hepes solution containing 100 mM methylamine (+MA). At high force, no additional steps are registered as it would be expected from a thioester bond cleavage event. Thereafter, we apply a protocol with subsequent pulses of decreasing mechanical load to investigate the force-dependency of the reaction. While at 30 pN no cleavage is observed, 100 s at 28 pN reveal one step that comes from the methylamine-induced cleavage of the thioester bond of one of the four Cpa domains (triangle). At 115 pN, the final extension of the molecule has increased by 45 nm, which originates from the polypeptide sequence released after thioester bond lysis. When held at 20 pN, the three remaining thioester bonds are cleaved (triangles; histogram inset #2: 38± 3.1 nm, mean±SD, n=21) and the final extension of the molecule increases for another 135 nm. b) Thioester bond cleavage probability as a function of force measured over a 100 s time-window. Data points are the mean and the error bars are the SD calculated using a jackknife analysis. The line represents a sigmoidal fit to the data (n=12 at 10 pN; n=20 at 15 pN; n=15 at 20 pN; n=9 at 21 pN; n=10 at 23 pN; n=9 at 24 pN; n=15 at 25 pN; n=15 at 27 pN; n=7 at 28 pN; n=15 at 30 pN; n=5 at 32 pN; n=6 at 35 pN). c) Rate of thioester bond cleavage as a function of force. Data points show the natural logarithm of the cleavage rate and the bars show the standard error of the mean. The curve represents a fit to the data described by a model that takes into account the effect of two sequential reactions: the rate of protein unfolding, which increases with force, and the rate of thioester bond cleavage, which decreases with the force. From this fit, we obtain a distance to the transition state for TED protein unfolding (x†U) of 0.9 nm, while the thioester bond cleavage exhibits a negative distance to the transition state (x†C = −0.4 nm), which suggests a requirement of a contraction of the Cpa polypeptide substrate to proceed with the cleavage of the bond, explaining its negative force-dependence. The dotted lines represent the individual unfolding and cleavage rates as obtained from the fit to the proposed model (Eq. S3, see Methods) (n=30 at 10 pN; n=38 at 15 pN; n=24 at 20 pN; n=23 at 21 pN; n=23 at 23 pN; n=24 at 24 pN; n=21 at 25 pN; n=37 at 27 pN; n=15 at 30 pN). Rate vs force dependency data was obtained in unrestricted time windows experiments.
