Figure 5. Cystamine-mediated abrogation of Cpa thioester bond reformation.
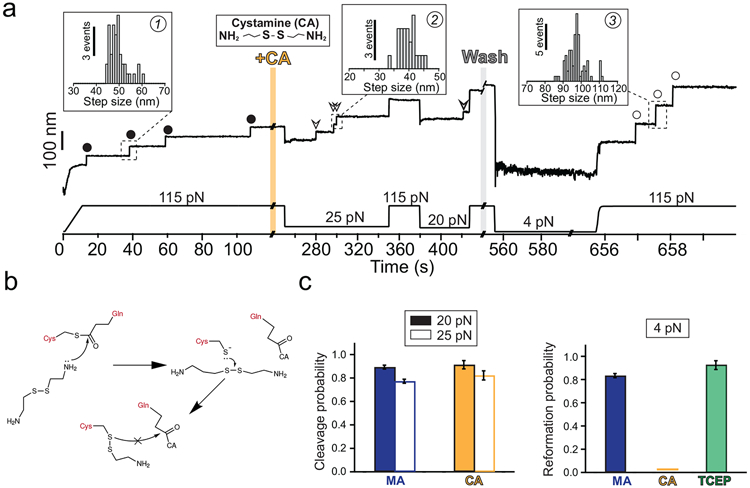
a) Magnetic tweezers trajectory of the Cpa polyprotein. After the unfolding of the thioester-intact Cpa domains at 115 pN (circles, inset histogram #1; 49.3 ± 3.8 nm, mean±SD, n=42), the buffer is exchanged and the polyprotein is exposed to a solution containing 100 mM cystamine (+CA). At 115 pN, no additional extensions are registered, but a drop in the force to 25 pN for 100 s leads to the appearance of three steps which account for the release of the polypeptide sequence trapped by the thioester bonds (empty arrows, inset histogram #2; 39.6 ± 2.7 nm for 25 pN, mean±SD, n=23). After the cleavage of all the bonds and after CA washout, force is quenched to 4 pN for 100 s to favor folding and reformation of the thioester. The final 115 pN pulse reveals three steps corresponding to thioester bond-cleaved Cpa domains (empty circles, inset histogram #3; 97.1 ± 5.2 nm, mean±SD, n= 78). b) Chemical scheme depicting the reformation blocking effect of CA. After the thioester bond nucleophilic cleavage by one of the CA primary amines, the free Cys thiol can attack CA disulfide bond (from the bound CA, or from another CA molecule). As a result, an intermolecular disulfide bond between Cpa Cys426 and CA is formed, preventing the thioester bond reformation. This disulfide reshuffling breaks the CA molecule and generates one free CA molecule (not shown in the scheme), and a Cys426-bound CA. c) Left panel compares the thioester bond cleavage probability by methylamine (MA) and CA at 20 and at 25 pN (MA, n=15 at 20 pN, n=15 at 25 pN; CA, n=8 at 20 pN, n=9 at 25 pN). Right panel compares the thioester bond reformation probability after 100 s at 4 pN after the treatment with MA, CA, and after the treatment with CA followed by TCEP (MA, n=16; CA, n=17; TCEP, n=6). Histogram bars are the mean and the error bars are the SD calculated using a jackknife analysis
