Figure 4.
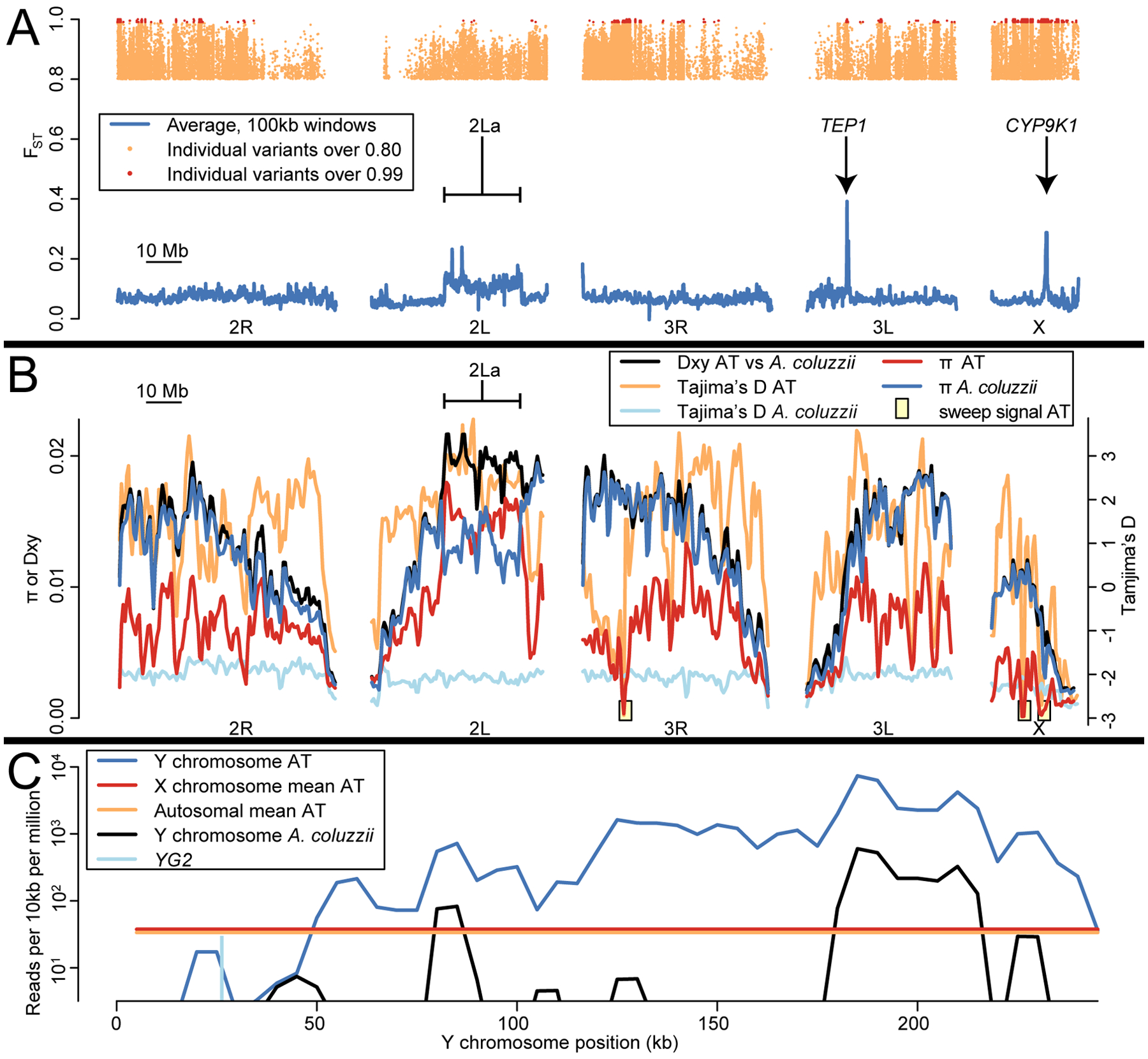
Unique genomic characteristics of AT. (A) FST across the genome between AT and Tengrela A. coluzzii. Tens of thousands of variants distributed across the genome are highly divergent between these taxa (FST > 0.8; orange dots at top), while over a thousand sites, concentrated in several clusters, are fixed or nearly so (“definitive differences”, FST > 0.99; red dots at top). Average FST in 100 kb windows is more modest (blue lines), but three regions stand out representing the 2La inversion, TEP1, and CYP9K1. (B) Nucleotide diversity (π), intertaxon divergence (Dxy), and site frequency spectra (Tajima’s D) in AT and A. coluzzii. Nucleotide diversity is low in AT except at the 2La inversion. Tajima’s D is mostly positive in AT, but three regions show low Tajima’s D, low π, and many high-FST sites (as shown in A), suggesting selective sweeps. (C) In most AT females, read coverage along most of the reference Y chromosome substantially exceeds the X/autosomal average. Coverage is negligible around sex-determining gene YG2. A. coluzzii females, in contrast, typically show negligible coverage across the entire Y except for a few repetitive sections.
