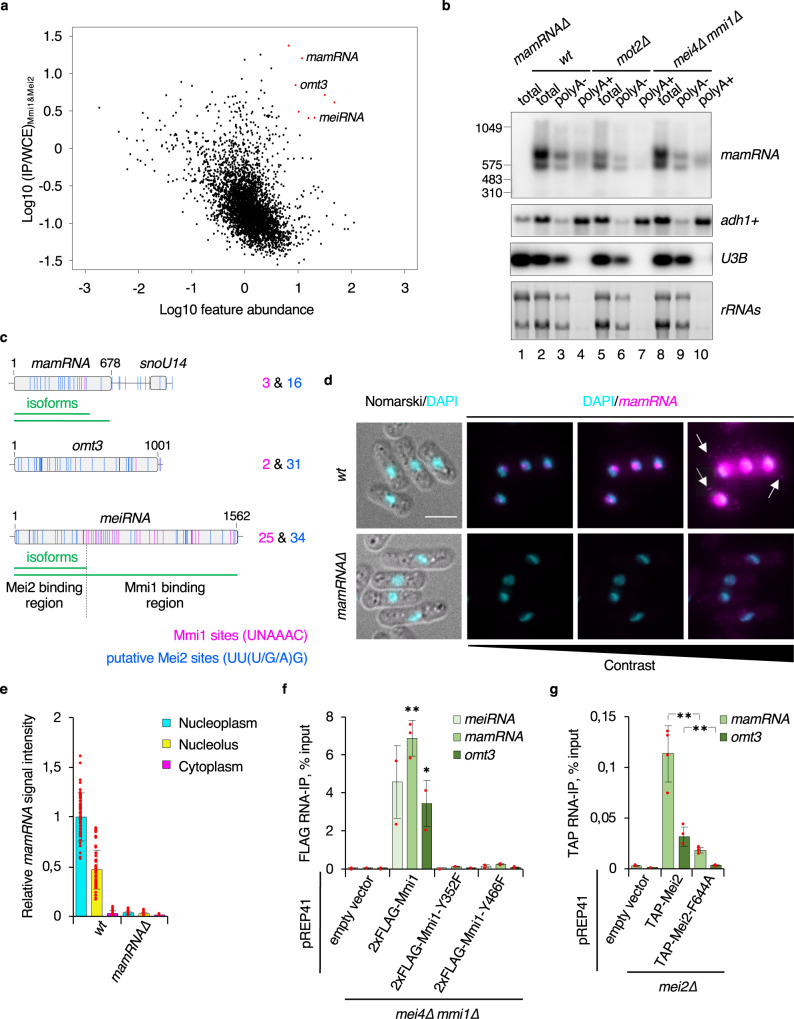
Fig. 2. Identification of mamRNA, an Mmi1 and Mei2-scaffolding lncRNA.
a SeqRIP-seq plot showing the enrichments (log10 IP/WCE) of transcripts pulled-down in the double-tagged strain of interest (y axis) as a function of their abundance (log10) (x axis) (n = 2). Red dots correspond to RNAs among the 100 most enriched and 100 most abundant features. Cells expressing TAP- or GFP-tagged Mmi1 and 3xFLAG-tagged Mei2 under the control of the mild nmt41 and strong nmt1 promoters, respectively, were used for immunoprecipitations (strain of interest: Pnmt41-TAP-Mmi1 Pnmt1-3xFLAG-Mei2; negative control: Pnmt41-GFP-Mmi1 Pnmt1-3xFLAG-Mei2). IP: Immunoprecipitate, WCE: Whole Cell Extract. b Northern blot showing mamRNA levels from total RNA samples and fractions enriched (polyA + ) or depleted (polyA-) for polyadenylated species, in the indicated genetic backgrounds. Enrichment and depletion of polyadenylated RNAs were verified by probing adh1 + mRNAs and snoU3B, respectively. Ribosomal RNAs serve as a loading control. c Scheme depicting the chromosomal loci encoding the mamRNA, omt3, and meiRNA lncRNAs with the number and position of Mmi1 (magenta) and putative non-overlapping Mei2 (blue) binding motifs. mamRNA and meiRNA isoforms are shown in green below the corresponding loci. The regions of Mmi1 and Mei2 binding to meiRNA are indicated35. d Representative images of mamRNA detected by smFISH in wt and mamRNA∆ mitotic cells. DNA was stained with DAPI. Images are shown as maximum-intensity projections of Z-stacks. Distinct contrast adjustments are shown to visualize mamRNA subcellular localization. White arrows point to cytoplasmic spots. The white scale bar represents 5 µm. e mamRNA signal intensities (integrated densities relative to nucleoplasmic average; mean ± SD; n = 50 cells) quantified from d in the indicated areas. f, g Enrichments (% input; mean ± SD; n = 3 or 4) of meiRNA, mamRNA and omt3 lncRNAs upon pulldown of wt or mutant 2xFLAG-tagged Mmi1 (f) and TAP-tagged Mei2 (g). Student’s t test (two-tailed) was used to calculate p-values. Between cells expressing pREP41-2xFLAG-Mmi1 and pREP41-2xFLAG-Mmi1-Y352F or Y466F (f), p = 0.00641 or 0.00663 (mamRNA) (0.01 > **>0.001); 0.03981 or 0.04065 (omt3) (0.05 > *>0.01). Between cells expressing pREP41-TAP-Mei2 and pREP41-TAP-Mei2-F644A (g), p = 0.00608 (mamRNA); 0.00916 (omt3) (0.01 > **>0.001). e, f, g Individual data points are represented by red circles.