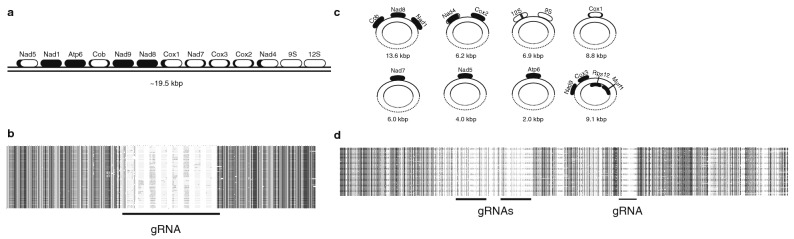
Figure 8.
Hypotheses for the structure and content of A. spiralis and P. ankaliazontas mitochondrial DNA. (a) A. spiralis ‘maxicircle’ DNA is likely linear, and approximately 19.5 kbp. A single contiguous DNA sequence could not be assembled; this hypothesis is based on consideration of long contigs from each of A. spiralis clones PhF-5 and PhF-6, along with manual correction of sequences. (b) A. spiralis PhF-6 gRNA-encoding maxicircles, also linear, are typically ~ 600 bp in length, and consist of a highly conserved region, along with a single gRNA-encoding region. A subset of 100 contigs was selected for display. (c) P. ankaliazontas encodes mitochondrial proteins on several ‘maxicircles’, most of which are circular-mapping. Each maxicircle has a conserved region. (d) P. ankaliazontas minicircles are also circular-mapping, are typically 1300–2200 bp, encode 2 gRNAs, and contain a conserved region. A subset of 100 contigs were selected for display. Black shading in genes in (a) and (c) correspond to a gene region that is edited; white regions are unedited. The frequency of U indel editing is presented for each gene in Supplementary Material, Table S2. Shading in (b) and (d) correspond to sequence identify of ≥ 90%, corresponding to minicircle backbone sequence, whereas divergent are similar to mitochondrial mRNAs, suggesting a role in RNA-editing.