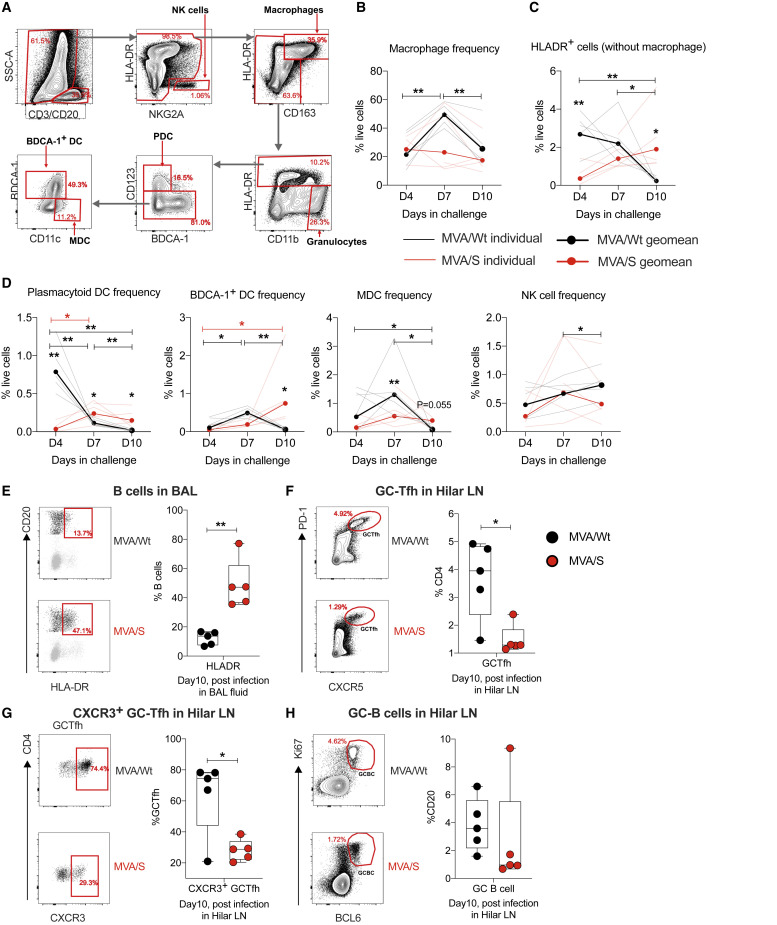
Figure 6.
Innate and adaptive immune responses after SARS-CoV-2 challenge in the lungs of rhesus macaques
Following SARS-CoV-2 infection, BAL fluid was collected on days 4, 7, and 10, and various innate cells were analyzed. On day 10 (at euthanasia), B cells were analyzed in BAL fluid, and GC-Tfh and GC-B cells were analyzed in hilar LNs. n =5/group, except for the MVA/S group on day 4, where data for one animal were not available. These analyses were performed only once because the macaque study was done only once.
(A) Gating strategy to identify innate cells in BAL fluid. Live cells were selected using a live/dead marker, and CD3+ and CD20+ cells were excluded. Then different innate cells were defined using the following combinations of markers: macrophages, HLA-DR+ CD163+; pDCs, HLA-DR+CD163−CD123+ BDCA1−; BDCA1+ DCs, HLA-DR+ CD163− CD123− BDCA1+; MDCs, HLA-DR+CD163−CD123−CD11C+; NK cells, HLA-DR− NKG2A+.
(B–D) Summary of changes in innate cell frequencies after infection.
(E) HLA-DR expression on B cells (CD20+) in BAL fluid on day 10 after infection. Left: representative flow plots. Right: summary of data for all animals.
(F–H) Total GC-Tfh (F), CXCR3+ GC-Tfh (G), and GC-B cells (H) on day10 after challenge in hilar LNs. Representative flow plots are shown on the right, and the data for all animals are shown on the left. GC-Tfh cells were CD3+CD4+CD8-CXCR5highPD-1high, and GC-B cells were CD3−CD20+BCL6+Ki67+.
NK, natural killer; pDC, plasmacytoid DC; MDC, myeloid-derived DC; LN, lymph node. Statistical differences between the groups and within the groups were analyzed using unpaired parametric t test and paired parametric t test, respectively. ∗p < 0.05, and ∗∗p < 0.005.