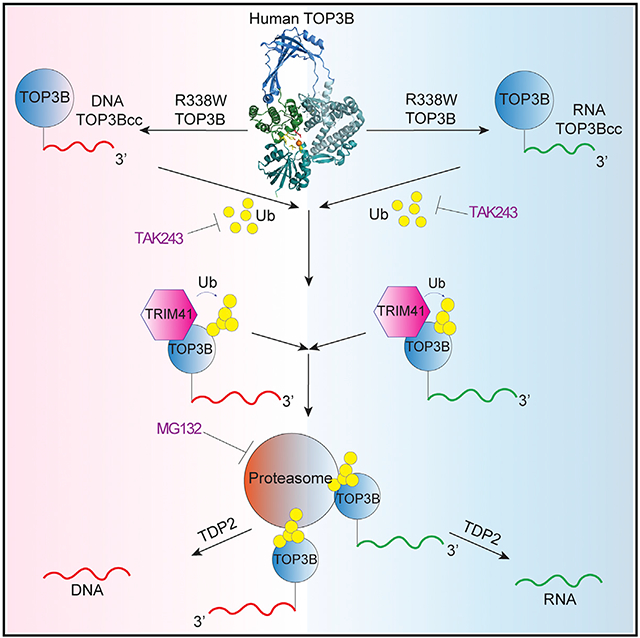
SUMMARY
The present study demonstrates that topoisomerase 3B (TOP3B) forms both RNA and DNA cleavage complexes (TOP3Bccs) in vivo and reveals a pathway for repairing TOP3Bccs. For inducing and detecting cellular TOP3Bccs, we engineer a “self-trapping” mutant of TOP3B (R338W-TOP3B). Transfection with R338W-TOP3B induces R-loops, genomic damage, and growth defect, which highlights the importance of TOP3Bcc repair mechanisms. To determine how cells repair TOP3Bccs, we deplete tyrosyl-DNA phosphodiesterases (TDP1 and TDP2). TDP2-deficient cells show elevated TOP3Bccs both in DNA and RNA. Conversely, overexpression of TDP2 lowers cellular TOP3Bccs. Using recombinant human TDP2, we demonstrate that TDP2 can process both denatured and proteolyzed TOP3Bccs. We also show that cellular TOP3Bccs are ubiquitinated by the E3 ligase TRIM41 before undergoing proteasomal processing and excision by TDP2.
Graphical Abstract
In Brief
Saha et al. introduce an approach to generate and detect the catalytic intermediates of TOP3B in DNA and RNA by engineering a self-poisoning enzyme, R338W-TOP3B. They reveal the cellular consequences of abortive TOP3Bcc formation and a repair pathway involving TRIM41, the proteasome, and TDP2 for processing of TOP3Bcc.
INTRODUCTION
Topoisomerases solve the topological constraints of nucleic acids during replication, transcription, recombination, chromosome segregation, and chromatin remodeling. They act by forming transient enzyme-nucleic acid intermediates by covalent phosphodiester bonds between their catalytic tyrosine residue and one end of the broken nucleic acid (3′ end for type IB and 5′ end for type IA and type II topoisomerases). These covalent catalytic intermediates are referred to as “topoisomerase cleavage complexes” (TOPccs). Normal TOPccs are readily reversible with resealing of the nucleic acid backbone after the topological changes and the release of topoisomerases for their next catalytic cycle (Pommier et al., 2016; Vos et al., 2011).
When TOPccs fail to reverse, eukaryotic cells use two tyrosyl DNA phosphodiesterases, namely, TDP1 and TDP2, to excise the 3′-tyrosyl-DNA and 5′-tyrosyl-DNA phosphodiester bonds involving TOP1 and TOP2, respectively (Cortes Ledesma et al., 2009; Pouliot et al., 1999). To gain access to those phosphotyrosyl bonds, TDPs require the prior degradation or denaturation of the covalently bound topoisomerases (Schellenberg et al., 2017; Sun et al., 2020b, 2020c). The ubiquitin-proteasome pathways play a pivotal role in the proteolysis of TOP1ccs and TOP2ccs, and the TDP excision pathways depend on ubiquitin-proteasome for TOP1cc and TOP2cc repair (Pommier et al., 2016; Sun et al., 2020b, 2020c). Despite extensive research on eukaryotic type IB and type II TOPccs, relatively little is known about eukaryotic type IA TOPccs formed by TOP3 enzymes.
Human topoisomerase 3B (TOP3B) remained remarkably understudied until the recent discovery that it is the only topoisomerase for both DNA and RNA (Ahmad et al., 2016, 2017a, 2017b; Stoll et al., 2013; Xu et al., 2013). Although TOP3B is not essential, Top3B knockout (KO) mice show autoimmunity (Kwan et al., 2007), infertility (Kwan et al., 2003), reduced lifespan (Kwan and Wang, 2001), abnormal neurodevelopment, and defective synapse formation (Ahmad et al., 2017a, 2017b; Xu et al., 2013). In humans, TOP3B genomic deletion has been linked with schizophrenia (Stoll et al., 2013) and autism spectrum disorders (ASDs) (Stoll et al., 2013; Xu et al., 2013) and observed in patients with autism, juvenile myoclonic epilepsy, cognitive impairment, facial dysmorphism, and behavior defects (Ahmad et al., 2017b; Daghsni et al., 2018; Kaufman et al., 2016). TOP3B genetic inactivation has also been linked with breast cancer, genomic instability, and renal cancer (Oliveira-Costa et al., 2010; Zhang et al., 2019).
TOP3B localizes to the nucleus and cytoplasm as it facilitates both DNA and RNA metabolic processes (Stoll et al., 2013). In the nucleus, TOP3B in association with the scaffolding protein TDRD3 (Tudor Domain Containing 3) is recruited to promoters to facilitate transcription by relaxing hypernegatively supercoiled DNA and resolving R-loops (Goto-Ito et al., 2017; Huang et al., 2018a; Siaw et al., 2016; Yang et al., 2014; Zhang et al., 2019). TDRD3 also associates with the mRNA-binding protein FMRP (Fragile X Mental Retardation Protein). Together, the three proteins form a large heterotrimeric complex (TOP3B-TDRD3-FMRP) associated with RNA during biogenesis and maturation (Lee et al., 2018; Stoll et al., 2013; Xu et al., 2013).
The present study introduces an approach to study TOP3Bccs in human cells. It provides evidence that TOP3Bccs are formed both in cellular DNA and RNA and presents insights about a molecular pathway for the cellular excision of abortive and cytotoxic TOP3Bccs.
RESULTS
Generation of Self-Trapping Mutant of TOP3B for In Vivo Detection of TOP3Bccs
To detect TOP3B cleavage complexes (TOP3Bccs), we generated a TOP3B mutant prone to remaining covalently linked to nucleic acids. Previous studies with Escherichia coli and Yersinia pestis topoisomerase I (Topo I) showed that substituting a single conserved arginine residue (Arg 321 in E. coli Topo I and Arg327 of Y. pestis Topo I) with a hydrophobic amino acid inhibits the resealing step of the enzyme catalytic cycle and stabilizes Topo I cleavage complexes (Narula et al., 2011). Sequence alignment (Figure 1A) revealed that Arg321 of E. coli and Arg327 of Y. pestis Topo I are conserved in human TOP3B (Arg338/R338). Hence, we hypothesized that mutating arginine (R338) to tryptophan might generate religation defective, self-trapping human TOP3B.
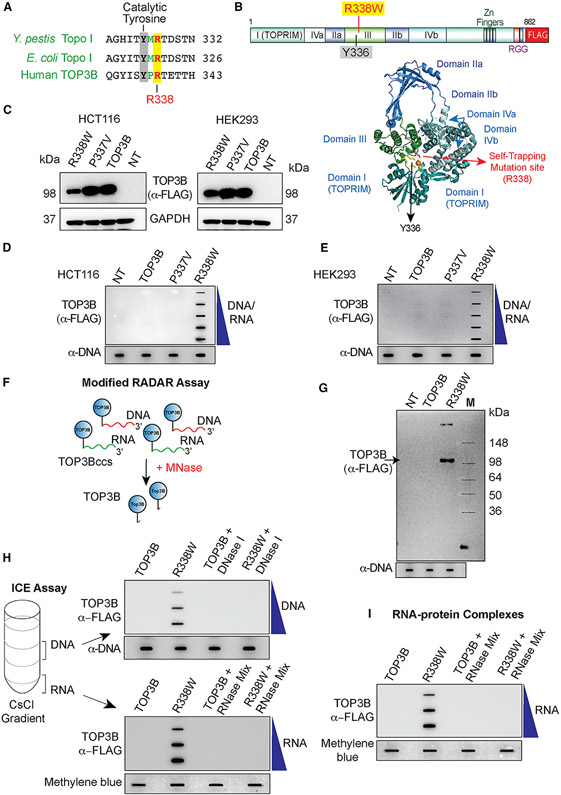
Figure 1. TOP3B Forms TOP3Bccs Both with DNA and RNA in Cells Transfected with R338W-TOP3B.
(A) Alignment of the active site regions of Y. pestis Topo I, E. coli Topo I, and human TOP3B.
(B) Structure of human TOP3B and ribbon representation of human TOP3B (amino acid [aa] residues 1–612) (Goto-Ito et al., 2017) with the active site Y336 and the self-trapping mutation site (R338).
(C) Ectopic expression of wild-type (WT) TOP3B, P337V, and R338W-TOP3B following transfection of HCT116 and HEK293 cells with the indicated TOP3B constructs for 72 h. Western blotting with anti-FLAG antibody.
(D and E) Detection of TOP3Bccs by RADAR assay in cells transfected with the indicated plasmid constructs for 72 h. TOP3Bccs were detected with anti-FLAG antibody. Equal loading was determined by slot blotting and probing with anti-dsDNA antibody. The figure is representative of three independent experiments. NT, mock-transfected cells.
(F) FLAG-tagged TOP3B (blue circles) cellular TOP3Bccs in DNA (red) and RNA (green) were digested with micrococcal nuclease (MNase) followed by SDS-PAGE and immunoblotting with anti-FLAG antibody.
(G) Modified RADAR assay in HEK293 cells transfected with WT or R338W-TOP3B for 72 h. TOP3B was detected with anti-FLAG antibody. Equal loading was tested by slot blotting and probing with anti-dsDNA antibody.
(H) TOP3Bccs in DNA and RNA of HEK293 cells transfected for 72 h with WT or R338W-TOP3B. Cesium chloride gradient ultracentrifugation was performed to separate DNA and RNA (middle and bottom of the gradient, respectively). DNA and RNA fractions were treated with excess RNase A (200 μg/mL) and RNase T1 (200 Units/ml) or DNase I (10 units) as indicated. DNA and RNA fractions were slot blotted and TOP3Bccs detected with anti-FLAG antibody. Equal loading was determined by slot blotting and probing with anti-dsDNA antibody (DNA) or methylene blue staining (RNA). The figure is representative of three independent experiments.
(I) TOP3Bccs in RNA of HEK293 cells transfected with WT or R338W-TOP3B for 72 h. Covalent protein-RNA adducts were isolated using TRIzol (Thermo Scientific) and treated with excess RNase A (200 μg/mL) and RNase T1 (200 units/ml) as indicated. Slot-blotted TOP3Bccs were detected with anti-FLAG antibody. Equal loading was determined by slot blotting and methylene blue staining. The figure is representative of three independent experiments.
See also Figure S1.
To test this hypothesis, we generated R338W-TOP3B by replacing the arginine 338 residue in the active site pocket of TOP3B (Figure 1B) with tryptophan and transfected human HCT116 and HEK293 cells with FLAG-tagged wild-type (WT)-TOP3B or R338W-TOP3B constructs. P337V-TOP3B was also included as control as the 337 residue is not conserved between bacterial (E. coli and Y. pestis) Topo I and human TOP3B (Figure 1A). After 3 days of transfection, WT-TOP3B, P337V-TOP3B, and R338W-TOP3B proteins were readily detectable by western blotting (Figure 1C). Transfected cells showed ≈25-fold overexpression for WT-TOP3B and ≈10-fold for R338W-TOP3B (Figures S1A and S1B). RADAR (rapid approach to DNA adduct recovery) assays (Kiianitsa and Maizels, 2013) to isolate nucleic acids with covalently bound protein adducts showed TOP3Bccs for R338W-TOP3B both in HEK293 and HCT116 cells (Figures 1D and 1E).
To confirm the specificity of the RADAR assay, nucleic-acid-containing protein adducts isolated by RADAR assay were digested with micrococcal nuclease (MNase) followed by SDS-PAGE electrophoresis and immunoblotting with anti-FLAG antibody (Figure 1F). Detection of a protein band corresponding to the size of human TOP3B (Figure 1G) indicated that R338W-TOP3B formed cellular TOP3Bccs.
TOP3B Forms TOP3Bccs In Vivo
To confirm the results obtained by RADAR assay and determine whether TOP3Bccs form both on DNA and RNA, we transfected HEK293 cells with FLAG-tagged TOP3B and R338W-TOP3B constructs and performed ICE (in vivo complex of enzymes) bioassays (Figure 1H). DNA- and RNA-protein adducts were separated from the free proteins by cesium chloride gradient ultracentrifugation. Slot blots of isolated DNA- and RNA-protein adducts showed that TOP3Bccs were present both in the DNA and RNA fractions (Figure 1H). In addition, the signals for DNA disappeared after DNase I and the RNA-associated signals disappeared after RNase treatment (Figure 1H).
To confirm this finding, we ectopically expressed R338W-TOP3B in HEK293 cells and isolated nucleic-acid-containing protein adducts by RADAR assay. Samples were digested either with excess RNase A and RNase T1 to remove RNA, with DNase I to digest DNA, or with MNase to remove both DNA and RNA (Figures S1C and S1D). TOP3Bcc signals were reduced but remained detectable after digestion with RNase A and RNase T1 mix or DNase I (Figures S1C and S1D) and eliminated by MNase, supporting our conclusion that R338W-TOP3B forms cellular TOP3Bccs both on DNA and RNA.
To further demonstrate the formation of TOP3Bccs on RNA, we isolated covalent RNA-protein adducts from cells transfected with R338W-TOP3B using TRIzol. Slot blotting confirmed the formation of cellular RNA-TOP3Bccs, which disappeared after RNase treatment (Figure 1I). Control ICE assays were also performed showing that TOP3B is the only eukaryotic topoisomerase forming RNA cleavage complexes, as TOP1 and TOP2 failed to form cleavage complexes in RNA under conditions where TOP1ccs and TOP2ccs were detected in cells treated with camptothecin or etoposide (Figures S1F and S1G).
TDP2 Excises Both DNA and RNA TOP3Bccs in Human Cells
In eukaryotes, irreversible TOP1ccs and TOP2ccs are processed by TDP1 and TDP2 (Sun et al., 2020b, 2020c). To determine whether TDP1 or TDP2 repair TOP3Bccs, we knocked down TDP1 or TDP2 or both using small interfering RNAs (siRNAs) (Figures S2A and S2B) in HEK293 cells and transfected FLAG-tagged R338W-TOP3B. RADAR assays showed that knocking down TDP2 increased TOP3Bccs, whereas TDP1 knockdown had no effect (Figures S2E and S2F).
To further establish the role for TDP2 in processing of TOP3Bccs, we repeated the experiments in isogenic TDP2KO HCT116 cells generated by CRISPR-Cas9 (Figure S2C; Huang et al., 2018b. TOP3Bccs were increased in TDP2KO cells (Figures 2A and 2B). These results indicate that TDP2 eliminates TOP3Bccs inside cells.
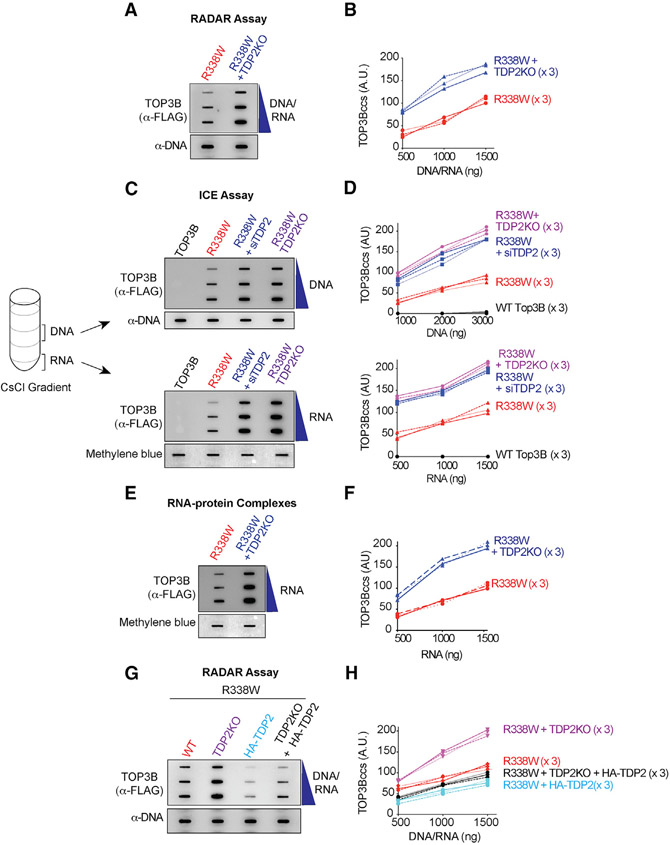
Figure 2. TDP2 Excises Cellular TOP3Bccs from Both DNA and RNA.
(A) WT or TDP2KO HCT116 cells were transfected with R338W-TOP3B. After 72 h, protein-nucleic acid adducts were isolated by RADAR assay. Slot-blotted TOP3Bccs were detected with anti-FLAG antibody. Loading was tested by slot blotting and probing with anti-dsDNA antibody.
(B) Quantitation of TOP3Bccs from three independent experiments as shown in (A). TOP3Bccs were measured by densitometric analyses of slot-blot signals and plotted as a function of total nucleic acid (DNA and RNA) concentration.
(C) HCT116 WT and TDP2KO cells were transfected with WT-TOP3B or R338W-TOP3B and siTDP2 constructs as indicated. After 72 h, ICE bioassays were performed to isolate DNA and RNA fractions. TOP3Bccs were detected with anti-FLAG antibody. Loading was tested with anti-dsDNA antibody or methylene blue staining (RNA).
(D) Quantitation of TOP3Bcc in three independent experiments as shown in (C).
(E) WT and TDP2KO HCT116 cells were transfected with FLAG-tagged R338W-TOP3B. After 72 h, protein-RNA adducts were isolated using TRIzol. TOP3Bccs were detected with anti-FLAG antibody. Loading was tested by methylene blue staining.
(F) Quantitation of TOP3Bcc in RNA in three independent experiments as shown in (E).
(G) Ectopic expression of TDP2 reduces TOP3Bccs. WT and TDP2KO HCT116 cells were transfected with FLAG-tagged R338W-TOP3B alone or co-transfected with HA-tagged TDP2. After 72 h, TOP3Bccs were detected with anti-FLAG antibody. Loading was tested with anti-dsDNA antibody.
(H) Quantitation of TOP3Bccs in three independent experiments as shown in (G).
See also Figure S2.
To determine whether TDP2 excises cellular TOP3Bcc from both DNA and RNA, we performed ICE assays. Knocking down TDP2 by siRNA in HEK293 cells enhanced TOP3Bccs both in the DNA and RNA fractions (Figures S2G and S2H). Similar results were obtained in the isogenic HCT116 TDP2KO cells (Figures 2C and 2D). To independently validate our finding, we took equal amounts of RADAR assay samples prepared from control and siTDP2-transfected HEK293 cells transiently expressing R338W-TOP3B and digested them either with excess RNase A and RNase T1 or with DNase I to distinguish the DNA and RNA TOP3Bccs. Downregulation of TDP2 increased both DNA and RNA TOP3Bccs (Figures S2K and S2L). Depletion of TDP2 also enhanced the RNA TOP3Bccs isolated with TRIzol from HEK293 cells transfected with R338W-TOP3B (Figures S2I and S2J). Consistent with this result, knocking out TDP2 in HCT116 cells enhanced TOP3Bccs in cellular RNA (Figures 2E and 2F).
Conversely, we performed TDP2 complementation experiments by transfecting hemagglutinin (HA)-tagged TDP2 (Figure S2D). TOP3Bccs were reduced both in WT and TDP2KO HCT116 cells ectopically expressing HA-tagged TDP2 (Figures 2G and 2H). Together, these results demonstrate the role of TDP2 in excising both cellular DNA and RNA TOP3Bccs.
TDP2 Processes Denatured or Proteolyzed but Not Native TOP3Bccs
To determine under which conditions TDP2 excises TOP3Bccs, we performed experiments with recombinant human TOP3B and recombinant TDP2 using an oligonucleotide substrate of sufficient length to detect TOP3Bccs by electrophoretic shift in SDS-PAGE (Figures 3A, 3B, and S3A). TOP3B produced a prominent TOP3Bcc (resulting in a retarded band in SDS-PAGE) (Figure 3B, lane 2). Less intense and slower migrating bands were also observed (Figure S3A, lane 2). To confirm that those bands correspond to different TOP3Bccs, we mapped the DNA cleavage sites associated with the TOP3Bccs by radiolabeling the oligonucleotide at the 5′ terminus and running DNA sequencing gels of the TOP3B reaction products (Figure S3B). Three cleavage sites were detected as 17-, 29-, and 55-nucleotide products. The most prominent site (17-nucleotide product) was at the tip of the hairpin loop structure (Figure S3B). To directly confirm that the upper-shifted bands were TOP3Bccs, we biotin-labeled the 3′ end of the oligonucleotide to detect covalently attached TOP3B (Figure S3C). Probing with streptavidin showed that TOP3B formed a prominent upper-shifted band and additional slower migrating bands with the biotin-labeled oligonucleotide. Probing with TOP3B antibodies confirmed that these bands corresponded to TOP3Bccs (Figure S3C).
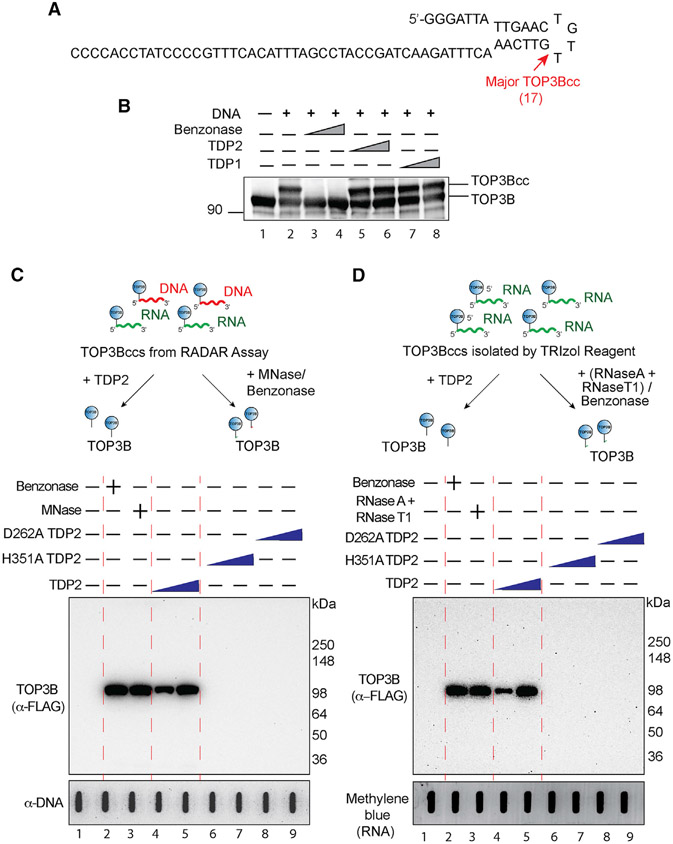
Figure 3. Recombinant Human TDP2 Excises Denatured but Not Native TOP3Bcc.
(A) Oligonucleotide substrate (69-mer) with main TOP3B site (17 nucleotides from the 5′ end).
(B) Recombinant human TDP2 does not excise native TOP3Bccs. The oligonucleotide (300 nM) was incubated with recombinant human TOP3B (4 μM). TOP3Bcc formation results in a slower migrating band (lane 2) (for full gel and additional minor bands see Figure S3A). TOP3Bccs were incubated with 1 or 3 μM recombinant TDP1 (lanes 7 and 8) or TDP2 (lanes 5 and 6). Benzonase (3 and 9 units, lanes 3 and 4,) was used as a positive control to degrade the oligonucleotide and release TOP3B. Samples were immunoblotted with anti-TOP3B antibody after SDS-PAGE.
(C) Recombinant human TDP2 excises denatured cellular DNA and RNA TOP3Bccs. HEK293 cells were transfected with FLAG-tagged R338W-TOP3B, and protein-nucleic acid adducts were recovered by RADAR assay. After incubation with recombinant TDP2 (3 and 6 μM, lanes 4 and 5), H351A TDP2 (3 and 6 μM, lanes 6 and 7), D262A TDP2 (3 and 6 μM, lanes 8 and 9), benzonase (250 units, lane 2), or MNase (300 units, lane 3), released TOP3B was detected by immunoblotting with anti-FLAG antibody after SDS-PAGE. Loading was tested by slot blotting and probing with anti-dsDNA antibody.
(D) Recombinant human TDP2 excises cellular RNA TOP3Bccs. HEK293 cells were transfected with R338W-TOP3B. Covalent protein-RNA adducts were isolated using TRIzol. After incubation with recombinant TDP2 (3 and 6 μM, lanes 4 and 5), H351A TDP2 (3 and 6 μM, lanes 6 and 7), D262A TDP2 (3 and 6 μM, lanes 8 and 9), benzonase (250 units, lane 2), or an excess amount of RNase A (200 μg/mL) and RNase T1 (200 units/ml) mix (lane 3), released TOP3B was detected by immunoblotting with anti-FLAG antibody after SDS-PAGE. Loading was tested by methylene blue staining (RNA).
See also Figure S3.
Having established biochemical conditions to generate TOP3Bccs, we studied the excision of TOP3Bccs by recombinant human TDP2. As expected, benzonase, which was used as a positive control, degraded the oligonucleotide and released free TOP3B (Figures 3B and S3A, lanes 3 and 4). However, neither TDP1 nor TDP2 processed the TOP3Bccs even at a high concentration (Figures 3B and S3A, lanes 5–8). From these experiments, we conclude that TDP2 cannot excise intact, native TOP3B from TOP3Bccs.
Next, we examined whether recombinant human TDP2 could process TOP3Bccs when TOP3B is denatured. To do so, we generated TOP3Bccs with the 3′-end biotin-labeled oligonucleotide substrate (Figure S3C) and found that ethanol-denatured TOP3Bccs were excised by TDP2, resulting in decreased TOP3Bcc band signal intensity (Figure S3D).
To further establish that TDP2 can excise denatured TOP3Bccs, we isolated TOP3Bccs by RADAR assay from R338W-TOP3B-transfected HEK293 cells. The RADAR assay uses a combination of chaotropic salt and detergent that denatures proteins and releases them from nucleic acids unless they are covalently attached to nucleic acids (Kiianitsa and Maizels, 2013). Those RADAR samples were incubated with increasing concentration of recombinant TDP2, catalytically defective TDP2 mutants (D262A and H351A TDP2) (Gao et al., 2012), MNase, or benzonase. Reaction mixtures were analyzed by SDS-PAGE followed by western blotting to detect released TOP3B. Untreated TOP3Bccs did not enter the gels due to their covalent linkage to nucleic acids (Figure 3C, lane 1). Benzonase and MNase, which were used as controls for complete degradation of the nucleic acids (DNA and RNA), released TOP3B detected as a single band corresponding to the size of FLAG-tagged TOP3B (~100 kDa) (Figure 3C, lanes 2 and 3). WT TDP2 also released TOP3B (Figure 3C, lanes 4 and 5). By contrast, the TDP2 catalytic mutants (D262A and H351A TDP2) failed to release TOP3B (Figure 3C, lanes 6–9). These results demonstrate that TDP2 can resolve TOP3Bccs when TOP3B is denatured.
We also studied the activity of TDP2 on proteolyzed TOP3Bccs by incubating TOP3Bccs digested with proteinase K (Figure S3E). TDP2 excised the proteolyzed TOP3Bccs by TDP2 ,as demonstrated by the release of the oligonucleotide substrate.
Next, we tested the processing of RNA TOP3Bccs by TDP2. We isolated the RNA TOP3Bccs from cells transfected with R338W-TOP3B by using the TRIzol procedure. Human recombinant TDP2, like benzonase or RNase A and RNase T1 mix, was able to release TOP3B from denatured RNA TOP3Bccs (Figure 3D). Together, these results demonstrate that TDP2 can excise denatured and proteolyzed but not native TOP3B from TOP3Bccs formed on DNA and RNA.
Ubiquitination and Proteasomal Processing of Cellular TOP3Bccs
To determine whether ubiquitin-mediated proteasomal degradation plays a role in the processing of TOP3Bccs, we treated HEK293 and HCT116 cells transfected with FLAG-tagged R338W or WT-TOP3B with the proteasome inhibitor MG132. Figures 4A, 4B, S4A, and S4B show that MG-132 increased TOP3Bccs both in HCT116 and HEK293 cells, implicating proteasomal degradation in the repair of cellular TOP3Bccs. We also treated HEK293 and HCT116 cells transfected with R338W-TOP3B with the E1 ubiquitin-activating enzyme (UAE) inhibitor TAK243 (Hyer et al., 2018) and found an enhancement of cellular TOP3Bccs by TAK243 (Figures 4C, 4D, S4C, and S4D). These results implicate proteasomal processing for the removal of TOP3Bccs.
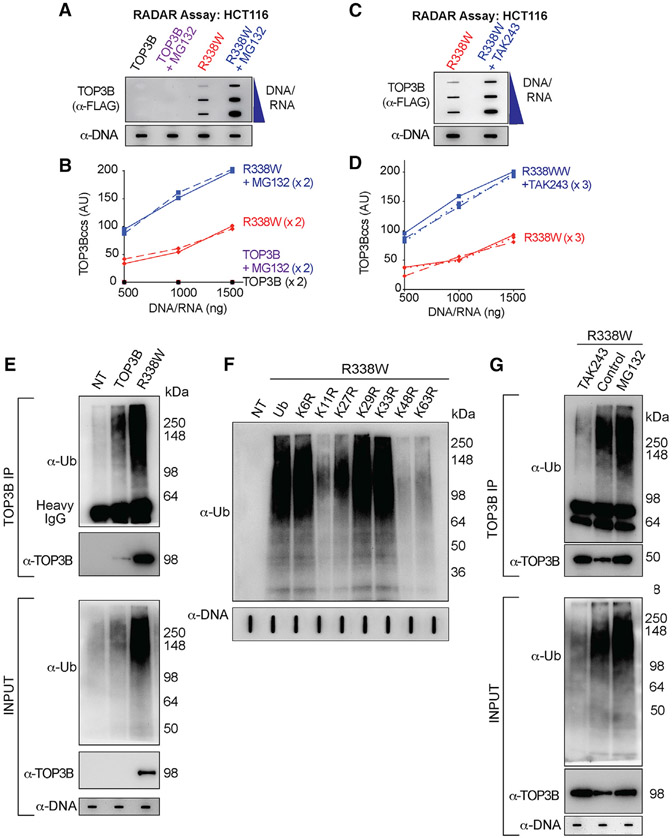
Figure 4. Cellular TOP3Bccs Are Ubiquitinated and Degraded by the Proteasomal Pathway.
(A) Proteasome inhibition enhances cellular TOP3Bccs. HCT116 cells were transfected with WT TOP3B and R338W-TOP3B for 72 h. Before harvest, cells were treated with MG132 (10 μM, 2 h). TOP3Bccs were detected by using anti-FLAG antibody. Loading was tested with anti-dsDNA antibody. The figure is representative of two independent experiments.
(B) Quantitation of TOP3Bcc in two independent experiments as shown in (A).
(C) Ubiquitination inhibition enhances cellular TOP3Bccs. HCT116 cells transfected with FLAG-tagged R338W-TOP3B for 72 h were treated with the UAE inhibitor TAK243 (10 μM, 2 h) before harvest. TOP3Bccs were detected with anti-FLAG antibody. Loading was tested by slot blotting and probing with anti-dsDNA antibody. The figure is representative of three independent experiments.
(D) Quantitation of TOP3Bcc in three independent experiments as shown in (C).
(E) Ubiquitination of cellular TOP3Bccs. RADAR assay samples were prepared from NT HCT116 cells or cells transfected with FLAG-tagged WT-TOP3B or R338W-TOP3B. Equal amounts of RADAR assay samples were immunoprecipitated (IPed) with anti-TOP3B antibody. IPed samples and RADAR samples were digested with MNase, resolved on SDS-PAGE, and immunoblotted with anti-Ub and anti-TOP3B antibodies. Loading (input) was tested with anti-dsDNA antibody.
(F) TOP3Bcc ubiquitination involves proteasomal-specific linkages to lysines K11, K27, K48, and K63. HCT116 cells were co-transfected with R338W-TOP3B and HA-tagged WT or mutant ubiquitin constructs for 72 h. RADAR samples were treated with MNase and probed with anti-Ub antibody. Loading was tested with anti-dsDNA antibody.
(G) Inhibition of TOP3Bcc ubiquitination by the UAE inhibitor TAK243 and enhancement by the proteasome inhibitor MG132. HCT116 cells transfected with FLAG-tagged R338W-TOP3B for 72 h were treated with either MG132 (10 μM, 2 h) or TAK243 (10 μM, 2 h), RADAR assay samples were isolated and IPed with an anti-TOP3B antibody. IP samples and the input RADAR assay samples were digested with MNase, resolved on SDS-PAGE and immunoblotted with anti-Ub and anti-TOP3B antibodies. Equal loading was tested by slot blotting and probing with anti-dsDNA antibody.
See also Figure S4.
To determine whether cellular TOP3Bccs are ubiquitinated, RADAR assay samples were immunoprecipitated (IPed) with anti-TOP3B antibody and digested with MNase to remove the DNA/RNA bound to TOP3Bccs. SDS-PAGE and immunoblotting with anti-ubiquitin antibody showed that cells transfected with R338W-TOP3B showed enhanced cellular ubiquitination of TOP3Bccs compared to WT-TOP3B-transfected cells (Figure 4E).
Ubiquitin (Ub) contains 7 lysine residues (K6, K11, K27, K29, K33, K48, and K63) for polyubiquitin chain formation. To determine the TOP3Bcc polyubiquitin linkages, we co-transfected HCT116 cells with R338W-TOP3B and either WT HA-tagged Ub or HA-tagged lysine-to-arginine Ub mutants for each of those 7 lysine residues (K6R-Ub, K11R-Ub, K27R-Ub, K29R-Ub, K33R-Ub, K48R-Ub, and K63R-Ub). K11, K27, K48, and K63 were critical for TOP3Bcc ubiquitination (Figure 4F), consistent with the proteasomal processing of TOP3Bccs.
To further establish the ubiquitination and proteasomal processing of TOP3Bccs, HCT116 cells were also transfected with R338W-TOP3B and treated with either MG132 or TAK243 before harvest. RADAR assay samples were prepared and IPed with the anti-TOP3B antibody and the levels of ubiquitinated TOP3Bccs determined as recently described for TOP1ccs and TOP2ccs (Sun et al., 2020a). While, as expected, MG132 increased TOP3Bcc ubiquitination, TAK243 reduced the ubiquitination of TOP3Bccs (Figure 4G). Together, these results show that the repair of trapped cellular TOP3Bccs is associated with their ubiquitination and proteasomal degradation.
TOP3Bccs Are Ubiquitinated by the E3 Ubiquitin Ligase TRIM41
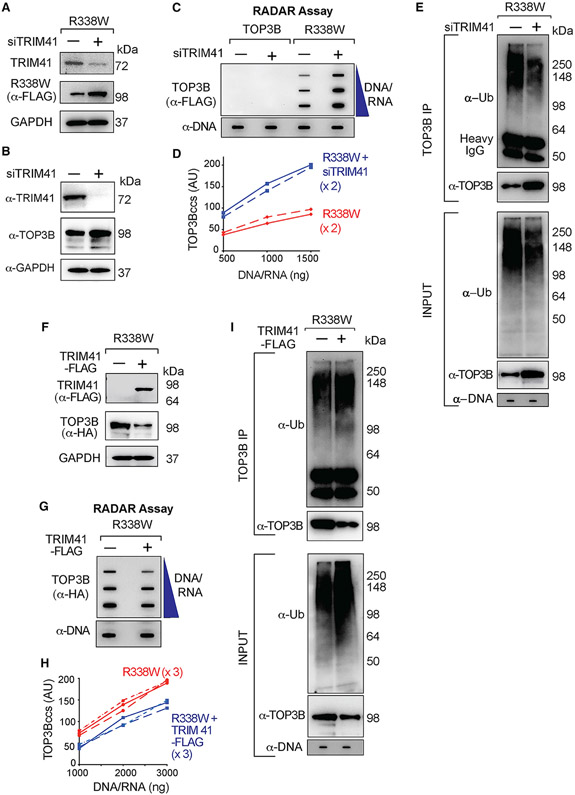
Because TRIM41 had been identified as an interaction partner of TOP3B by the yeast 2-hybrid assay (Kobayashi and Hanai, 2001) and high-throughput proteome analyses (Rolland et al., 2014), we tested whether TRIM41 is an E3 ligase for TOP3Bccs. Cells transfected with siTRIM41 accumulated more R338W-TOP3Bccs and showed elevated TOP3B (Figures 5A-5D). This finding was confirmed with different siRNA against TRIM41 (Figures S5B and S5C). Immunoprecipitation (IP) of the RADAR samples with anti-TOP3B showed that downregulation of TRIM41 reduced ubiquitinated TOP3Bccs (Figure 5E).
Figure 5. TRIM41 Acts as a Ubiquitin Ligase for TOP3Bccs and Promotes the Repair of TOP3Bccs.
(A) Immunoblots showing TRIM41 and R338W-TOP3B expression after TRIM41 downregulation (GAPDH as loading control). HCT116 cells were either transfected with a FLAG-tagged R338W-TOP3B plasmid construct alone or co-transfected with a siTRIM41construct for 72 h.
(B) Immunoblots showing effect of TRIM41 depletion on endogenous TOP3B level. GAPDH was included as loading control. HCT116 cells were transfected with siTRIM41construct for 72 h.
(C) HCT116 cells were transfected with FLAG-tagged WT- or R338W-TOP3Bs or co-transfected with siTRIM41construct for 72 h. TOP3Bcc were detected with anti-FLAG antibody. Equal loading was tested with anti-dsDNA antibody. The figure is representative of two independent experiments.
(D) Quantitation of TOP3Bccs in two independent RADAR assays as shown in (C).
(E) HCT116 cells were transfected with FLAG-tagged R338W-TOP3B alone or with siTRIM41 construct. After 72 h, equal amounts of RADAR assay samples were IPed with anti-TOP3B antibody. IP samples and the input RADAR assay samples were digested with MNase, resolved on SDS-PAGE and immunoblotted with anti-Ub and anti-TOP3B antibodies. Loading of input RADAR samples was tested with anti-dsDNA antibody.
(F) Immunoblots showing TRIM41 and R338W-TOP3B expression after transfection of HCT116 cells with HA-tagged R338W-TOP3B plasmid construct alone or with FLAG-tagged TRIM41. GAPDH served as loading control.
(G) Reduced TOP3Bccs upon TRIM41 overexpression. HCT116 cells were transfected with HA-tagged R338W-TOP3B alone or co-transfected with FLAG-tagged TRIM41 for 48 h. TOP3Bccs were detected with anti-HA antibody. Loading was tested by with anti-dsDNA antibody. The figure is representative of three independent experiments.
(H) Quantitation of TOP3Bcc formation in three independent RADAR assays as shown in (F).
(I) Increased TOP3Bcc ubiquitination upon transfection with TRIM41. HCT116 cells were either transfected with HA-tagged R338W-TOP3B alone or co-transfected with FLAG-tagged TRIM41. After 48 h, RADAR assay samples were IPed with anti-TOP3B antibody. IP samples and the input RADAR assay samples were digested with MNase, resolved on SDS-PAGE, and immunoblotted with anti-Ub and anti-TOP3B antibodies. Loading was tested with anti-dsDNA antibody.
See also Figure S5.
To further test the implication of TRIM41 in the repair of TOP3Bccs, we overexpressed TRIM41 (Figure 5F) in HCT116 cells transfected with R338W-TOP3B. Overexpression of TRIM41 decreased cellular TOP3Bccs (Figures 5G and 5H), and IP of the RADAR samples with anti-TOP3B antibody showed increased ubiquitinated TOP3Bccs (Figure 5I). These results implicate TRIM41 in the ubiquitination and repair of TOP3Bccs.
We repeated those experiments with a TRIM41 RING domain mutant lacking the E3 ubiquitin ligase activity (deletion of amino acids 16–60; ΔRING-TRIM41) (Figure S5D; Lassot et al., 2018; Patil et al., 2020). Transfection of WT-TRIM41 lowered R338W-TOP3Bccs, whereas transfection of the ΔRING-TRIM41 failed to decrease TOP3Bccs (Figures S5E and S5F). In parallel, WT-TRIM41 increased the ubiquitination of TOP3Bccs, whereas the ΔRING-TRIM41 mutant did not (Figure S5G). Combined, these results show that the E3 ubiquitin ligase activity of TRIM41 is required for the ubiquitylation and removal of TOP3Bccs.
TRIM41-Mediated Ubiquitination and Proteasomal Processing of TOP3Bccs Are Coordinated with the Excision of TOP3Bcc by TDP2
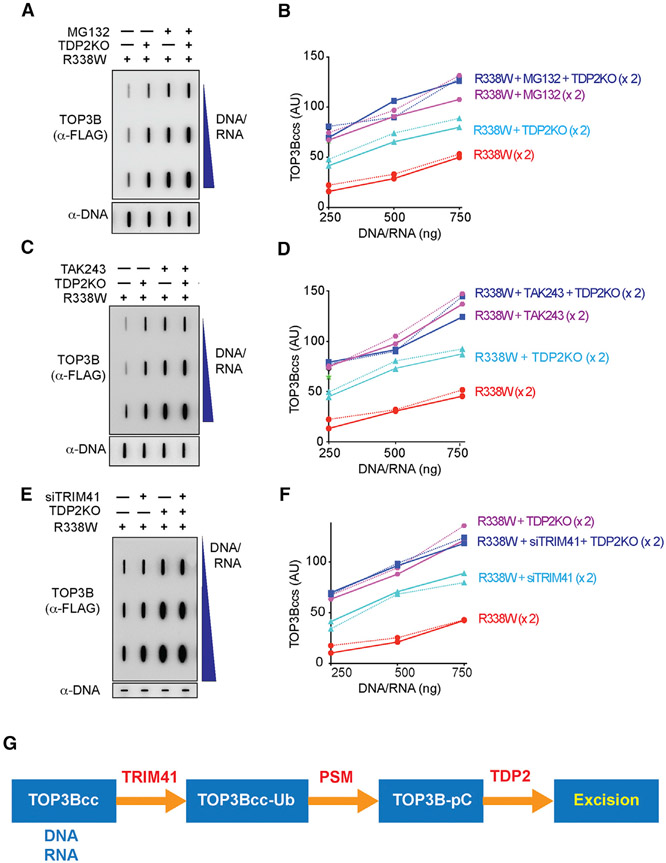
Next, we investigated the relationship between the processing of TOP3Bccs by TDP2 and the ubiquitin-proteasome pathway in WT and TDP2KO HCT116 cells transfected with R338W-TOP3B. Proteasomal inhibition by MG-132 produced a greater accumulation of TOP3Bccs than TDP2 inactivation (Figures 6A and 6B). Combining both produced no further increase in cellular TOP3Bccs compared to MG132 single treatment. We repeated those experiments with TAK243 and found that knocking out TDP2 did not increase TOP3Bcc levels further in TAK243-treated cells (Figures 6C and 6D). These results are consistent with the proteasomal processing of ubiquitinated TOP3Bccs prior to TDP2 activity.
Figure 6. TDP2-Mediated Repair of TOP3Bccs Is Dependent on Ubiquitination and Proteasomal Processing.
(A) WT and TDP2KO HCT116 cells were transfected with FLAG-tagged R338W-TOP3B alone and incubated for 72 h. Before harvest, cells were treated with MG132 (10 μM, 2 h) as indicated. Nucleic acids and protein-nucleic acid adducts were recovered by RADAR assay. TOP3Bccs were detected using anti-FLAG antibody. Equal loading was with anti-dsDNA antibody. The figure is representative of two independent experiments.
(B) Quantitation of TOP3Bccs in two independent experiments as shown in (A).
(C) WT and TDP2KO HCT116 cells were transfected with FLAG-tagged R338W-TOP3B alone and incubated for 72 h. Before harvest, cells were treated with TAK243 (10 μM, 2 h). Nucleic acids and protein-nucleic acid adducts were recovered by RADAR assay, and TOP3Bccs were detected using anti-FLAG antibody. Equal loading was tested with anti-dsDNA antibody. The figure is representative of two independent experiments.
(D) Quantitation of TOP3Bccs in two independent RADAR assays as shown in (C).
(E) WT and TDP2KO HCT116 cells were transfected with FLAG-tagged R338W-TOP3B alone or co-transfected with siTRIM41 constructs and incubated for 72 h. TOP3Bccs were detected after RADAR assay with anti-FLAG antibody. Equal loading was tested with anti-dsDNA antibody. The figure is representative of two independent experiments.
(F) Quantitation of TOP3Bccs in two independent RADAR assays as shown in (E).
(G) Model for the processing of TOP3Bccs by TRIM14, the proteasome (PSM), and TDP2. See also Figure S6.
We also tested whether TRIM41 affected TDP2-mediated processing of TOP3Bccs. WT and TDP2KO HCT116 cells were transfected with FLAG-tagged R338W-TOP3B alone or co-transfected with siTRIM41 constructs. Compared to siTRIM41-treated cells, TDP2KO cells showed enhanced accumulation of TOP3Bccs. In addition, TRIM41 depletion in TDP2KO cells did not show further increase in TOP3Bcc levels (Figures 6E and 6F). These results are consistent with TRIM41 and TDP2 acting in the same repair pathway for TOP3Bccs excision (Figure 6G).
In parallel, we examined whether inactivating TDP2, TRIM41, or the proteasome affected TOP3Bccs generated by transfection of WT-TOP3B or endogenous TOP3B (Figure S6). To detect endogenous TOP3Bccs and/or the TOP3Bccs generated by transfection of WT-TOP3B, we IPed TOP3Bccs from RADAR assay samples. Endogenous TOP3Bcc were not detectable following IP enrichment in cells lacking TDP2 or TRIM41; however, they were detectable after blocking the ubiquitin-proteasomal pathway with MG132 or TAK243 (Figure S6A). For WT-TOP3B-transfected cells, TOP3Bccs were detectable after IP with TOP3B antibody in cells expressing TDP2 and TRIM41, and MG132 or TAK243 increased TOP3Bccs further (Figure S6B). We conclude that TRIM41-mediated ubiquitination and proteasomal processing of TOP3Bccs are critical for the excision of TOP3B by TDP2 (Figure 6G).
Phenotypic Consequences of Trapping TOP3B in Human Cells
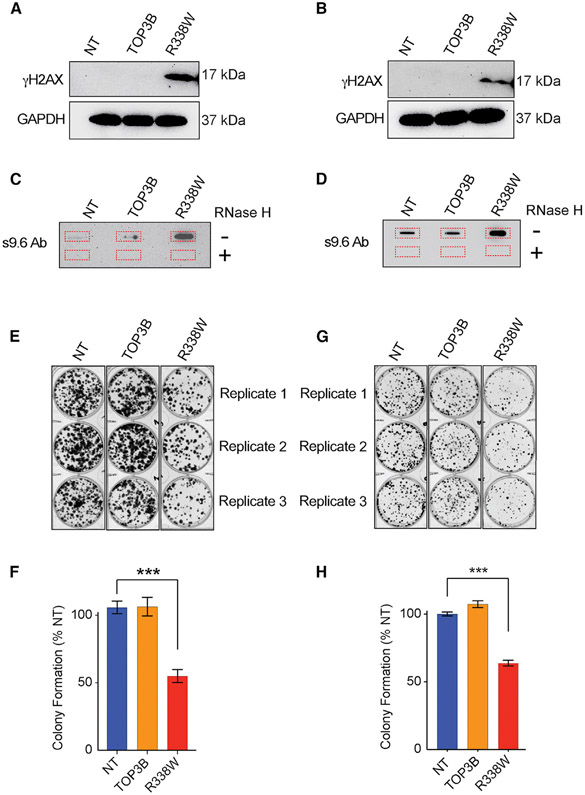
To explore the cellular effects of TOP3Bccs, we transfected HCT116 and HEK293 cells with the self-poisoning R338W-TOP3B and measured histone γH2AX induction, R-loop formation, and cell growth (Figures 7). R338W-TOP3B produced γH2AX signal both in HEK293 and HCT116 cells, indicating that trapped TOP3Bccs cause genomic DNA damage (Figures 7A and 7B).
Figure 7. Cellular Consequences of TOP3B Trapping and Identification of RNAs Associated with TOP3B.
(A) Trapping of TOP3B results in γH2AX induction. Immunoblots showing γH2AX levels in HEK293 cells transfected with R338W-TOP3B, WT-TOP3B, or NT.
(B) Trapping of TOP3B results in γH2AX induction in HCT116 cells transfected with R338W-TOP3B, WT-TOP3B, or mock-transfection reagent (NT).
(C) Trapping of TOP3B results in R-loop accumulation. Genomic DNA isolated from HEK293 cells transfected with R338W-TOP3B, WT-TOP3B, or NT was slot blotted and probed with S9.6 antibody.
(D) Trapping of TOP3B results in R-loop accumulation in HCT116 cells transfected with R338W-TOP3B, WT-TOP3B, or NT.
(E) Representative images of colony formation assay in HEK293 cells after transfection with R338W-TOP3B, WT-TOP3B or NT.
(F) Quantitative representation of colony formation assays as shown in (E). Data are provided as means ± standard deviations (SD) (n = 3). ***p ≤ 0.0005 (two-tailed unpaired t test).
(G) Representative images of colony formation assay in HCT116 cells after transfection with R338W-TOP3B, WT-TOP3B, or NT.
(H) Quantitative representation of colony formation assays as shown in (G). Data are provided as means ± standard deviations (SD) (n = 3). ***p ≤ 0.0005 (two-tailed unpaired t test).
Because TOP3B has been proposed to suppress R-loops (Huang et al., 2018a; Wilson-Sali and Hsieh, 2002; Yang et al., 2014; Zhang et al., 2019), we performed slot blotting of genomic DNA with the S9.6 antibody. Compared to control (mock)-transfected (NT) cells and WT-TOP3B-transfected cells, R338W-TOP3B-transfected cells (both HEK293 and HCT116) displayed higher R-loop levels (Figures 7C and 7D). The S9.6 signal was suppressed after RNase H treatment, which is indicative of R-loops. We conclude that trapping TOP3Bccs induces R-loops.
Compared to NT and WT-TOP3B-transfected cells, R338W-TOP3B-transfected cells also showed reduced colony formation capacity (Figures 7E, 7F, 7G, and 7H). These results are consistent with the potentially deleterious genomic consequences of TOP3B trapping.
DISCUSSION
Our study addresses two outstanding questions: (1) how to detect and induce cellular TOP3Bccs and (2) how cells repair TOP3Bccs in DNA and RNA. Using a self-poisoning TOP3B (R338W-TOP3B), we show that TOP3Bccs form both in RNA and DNA in human cells. We reveal a pathway for the excision of TOP3Bccs consisting in the sequential action of TRIM41 as E3 ubiquitin ligase and the proteasome giving access for TDP2 to excise TOP3Bccs. Using R338W-TOP3B, we show the deleterious consequences of TOP3Bcc accumulation (γH2AX and R-loop accumulation and reduced cell growth).
The catalytic cycle of topoisomerases consists in self-reversible and normally transient cleavage complexes (TOPccs). To capture TOPccs formed by type IB and IIA topoisomerases, small molecules are used as probes and therapeutic antitumor and antibacterial agents (Maxwell, 1999; Nitiss, 2009; Pommier et al., 2016). For type IA topoisomerases, there is no known small-molecule “topoisomerase poison.” However, screening recombinant Y. pestis Topo I for SOS-inducing mutants identified point mutations in the TOPRIM motif and active site pocket of the enzyme that produce TOPccs (Cheng et al., 2005, 2009; Narula et al., 2011). Biochemical studies with the Arg321Trp mutant of E. coli Topo I showed defective DNA rejoining of the TOPccs as well as a partial defect in DNA cleavage (Narula et al., 2011). This defect was explained by the fact that the positively charged conserved arginine residue, which is adjacent to the active site tyrosine and divalent magnesium ion, is critical for the alignment and nucleophilic attack of the topoisomerase phosphotyrosyl bond by the 3′-hydroxyl-end of the cleaved nucleic acid (Narula et al., 2011). Our study shows that the corresponding arginine of TOP3B is also critical for the reversal of TOP3Bccs both in DNA and RNA, as substitution to tryptophan (R338W) produces a potent self-trapping TOP3B.
To excise abortive and potentially damaging TOPccs, eukaryotic cells use two different tyrosyl DNA-phosphodiesterases, namely, TDP1 and TDP2 (Sun et al., 2020b, 2020c). In vitro and in vivo properties of TDP2 made us hypothesize that TDP2 might resolve TOP3Bccs. Indeed, purified human TDP2, apart from processing double-stranded 5′-tyrosyl overhang substrates (mimicking TOP2ccs), is most active with single-stranded DNA substrates bearing a 5′-phosphotyrosine terminus (mimicking DNA-TOP3Bccs) (Ahmad et al., 2017a; Gao et al., 2012). Recombinant human TDP2 also has the ability to process a 5′ tyrosine covalently linked to a ribonucleotide or a polyribonucleotide (mimicking RNA-TOP3Bccs), and the co-crystal structure of TDP2 has shown its molecular interaction with a tyrosyl-RNA substrate (Gao et al., 2014). Finally, TDP2 is known to act as the VPg unlinkase that hydrolyzes the covalent phosphotyrosyl bond between the viral VPg protein and the 5′ end of viral RNAs (mimics of RNA-TOP3Bccs) during the replication of picornaviruses (Kawale and Povirk, 2018; Pommier et al., 2014; Virgen-Slane et al., 2012). Consistent with the cellular role of TDP2 for excising TOP3Bccs, we found that both DNA and RNA TOP3Bccs accumulate in TDP2-depleted cells.
Polyubiquitin chains form through one or more of the seven internal lysines of ubiquitin (Hatakeyama, 2017), and we found that TOP3Bccs are preferentially modified on K48, K11, and K63 and to a lesser extent on K27. K48-linked polyubiquitin chains mainly target substrate proteins for proteasomal degradation (Chau et al., 1989; Johnson et al., 1995). Polyubiquitin chains formed by K11 and K63 can also promote proteasomal targeting (Bedford et al., 2011; Kravtsova-Ivantsiv and Ciechanover, 2012; Meyer and Rape, 2014; Saeki et al., 2009). In addition, K27 or K63 polyubiquitin chains may help in docking TDP2 to TOP3Bccs and stimulating TDP2 by engaging its UBA domain as it does for TOP2ccs (Schellenberg et al., 2020). Overall, our results establish the importance of the ubiquitin-proteasomal pathway for the repair of TOP3Bccs in addition to its role for both TOP1ccs and TOP2ccs (Sun et al., 2020b, 2020c).
We demonstrate that TRIM41 (also known as RINCK) acts as E3 ubiquitin ligase for TOP3Bccs. TRIM41 is a member of the tripartite motif (TRIM) protein family, which is one of the largest families of the single-protein RING-type E3 ubiquitin ligases (Hatakeyama, 2017; Meroni and Diez-Roux, 2005; Napolitano and Meroni, 2012; Tanaka et al., 2005). TRIM41 is a bona fide E3 ubiquitin ligase mediating ubiquitination and degradation of different substrates, including protein kinase C (Chen et al., 2007) and the transcription factor ZSCAN21 (Lassot et al., 2018). Identification of TRIM41 as a binding partner of TOP3B was previously reported (Kobayashi and Hanai, 2001; Rolland et al., 2014) but had not been connected to its role as an E3 ubiquitin ligase. Experimental evidence provided by our study further shows that the RING domain of TRIM41 is important for the repair of TOP3Bccs.
The fact that TDP2 depletion increases TOP3Bccs suggests that the proteasome is insufficient to remove TOP3Bcc in the absence of TDP2. The high levels of TOP3Bccs following transfection with R338W-TOP3B might saturate the proteasome or TDP2 may act as a rate-limiting step for full proteasomal processing of trapped TOP3B. Similarly, earlier publications show that TDP1 or TDP2 depletion increases TOP1ccs and TOP2ccs. Specifically, TDP1 depletion increases TOP1ccs in Tdp1−/− neural tissue (Katyal et al., 2014) and TOP1MTccs in mitochondria (Chiang et al., 2017; Ghosh et al., 2019). Two additional publications show that TDP2−/− cells accumulate more TOP2ccs (Hoa et al., 2016; Sasanuma et al., 2018). We cannot exclude the possibility that TDP2 may also act on TOP3Bccs independently of proteolysis. This would require denaturation of TOP3Bccs, as shown in our biochemical experiments. Such a scenario has been recently observed for the processing of TOP2cc by TDP2 (Schellenberg et al., 2017) and remains hypothetical for TOP3B. But, our biochemical assays indicate that TDP2 cannot excise TOP3Bccs unless TOP3B is denatured or proteolyzed (Figures 3 and S3). This finding is in line with the fact that purified TDP2 is unable to process native TOP2ccs in vitro unless TOP2B is proteolyzed or denatured (Gao et al., 2014; Schellenberg et al., 2017).
In conclusion, our study provides direct evidence for the activity of TOP3B on cellular DNA and RNA. It demonstrates that TDP2 excises not only TOP2 and TOP1 cleavage complexes but also TOP3Bccs both in DNA and RNA. It reveals the role of TRIM41 as an E3 ubiquitin ligase for TOP3Bccs. Based on the redundancy of the repair pathways for TOP1 and TOP2 cleavage complexes (Sun et al., 2020a, 2020b, 2020c), further studies are warranted to determine whether additional ubiquitylation, proteolytic, and endonucleolytic pathways are involved in the repair of TOP3Bccs. Finally, the self-poisoning mutant R338W-TOP3B is likely to provide a molecular tool to study and map TOP3Bccs in DNA and RNA.
STAR★METHODS
RESOURCE AVAILABILITY
Detailed methods are provided in the online version of this paper and include the following:
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Lead Contact Yves Pommier (pommier@nih.gov).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
Imaging data supporting the current study have been deposited at Mendeley Data (https://doi.org/10.17632/jp9vs3sddy.1).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HEK293 (ATCC, Manassas, VA) and HCT116 (Developmental Therapeutics Program, National Cancer Institute) cell lines were grown in Dulbecco’s modified Eagle’s medium (Life Technologies, Carlsbad, CA) supplemented with 10% Fetal Bovine Serum (Gemini, West Sacramento, CA) and 1% penicillin-streptomycin (ThermoFisher Scientific, 15140122) at 37°C in humidified 5% CO2 chamber. HCT116 TDP2 knockout (TDP2KO) cells were generated by CRISPR/Cas9 method as previously described (Huang et al., 2018b). Tni-FNL cells were cultured in GIBCO Express 5 medium with 18mM glucose.
METHOD DETAILS
Mammalian Expression Constructs and Transient Expression in Mammalian Cell
Human TOP3B-Myc-FLAG cDNA ORF (CAT#: RC223204) and Human TRIM41-Myc-FLAG cDNA ORF (CAT#: RC210557) Clones were purchased from OriGene. The full-length cDNAs of TDP2 and TOP3B were PCR-amplified from human pCMV6-AN-DDK(Flag)-hTDP2 ORF clone (Huang et al., 2018b) and TOP3B-Myc-FLAG cDNA ORF Clone (CAT#: RC223204) respectively using cloning primers (TDP2 forward primer 5′- TATAGGATCCGAGTTGGGG AGTTGCCTG-3′; TDP2 reverse primer 3′- GCGCGAATTCTTACAATATTATATCTAA-5′; TOP3B forward primer 5′- GCTTGGATCCAAGACTGTGCTCATGGTT-3′; TOP3B reverse primer 3′- CCAAGAATTC TCATACAAAGTAGGCGGC-5′) and subcloned into pcDNA3-HA with BamHI and EcoRI sites. HA-Ubiquitin WT plasmid was a gift from Ted Dawson (Addgene plasmid CAT#: 17608). Plasmids were transfected in HCT116 and HEK293 cells using Lipofectamine 3000 Reagent (CAT#: L3000015, ThermoFisher Scientific) according to the manufacturer’s protocol for 48–72 h.
Recombinant Human TOP3B Production
TOP3B was initially PCR amplified from Human TOP3B-Myc-FLAG cDNA ORF (CAT#: RC223204) using forward primer: 5′-CGGGGTACCATGAAGACTGTGCTCATGG-3′ and reverse primer: 5′-CCGCTCGAGTCATACAAAGTAGGCGGCCAG-3′ and cloned into Gateway entry vector pENTR3C (Invitrogen, CAT#: A10464). TOP3B was then subcloned by Gateway LR recombination (Thermo Fisher) into pDest-635 (22876-X01-635) for insect cell expression which includes an N-terminal His6 tag. Bacmid was prepared in DE77, a DH10Bac-derived strain (Bac-to-Bac system, Thermo Fisher) and after purification, bacmid DNA was verified by PCR amplification across the bacmid junctions. Bacmids were transfected in SF-9 cells using PEI (1 mg/ml with 5% glucose; Polysciences, CAT#: 23966), recombinant baculovirus stock was collected and titrated using ViroCyt (Beckamn). Two liters of Tni-FNL cells were set in a baffled 5-l Thomson Optimum Growth Flask in GIBCO Express 5 medium with 18mM glucose at a cell density of 1 × 106 cells/ml at 27°C and 24 hr later infected at a MOI (multiplicity of infection) of 3. After 3 days of incubation at 21 °C, cell pellets were collected by centrifugation at 2000 rpm for 11 min and flash frozen on dry ice. Cell pellet was thawed by the addition of 200 mL of lysis buffer (20 mM HEPES, 300 mM NaCl, 1 mM TCEP and 1:100 v/v of Sigma protease inhibitor P8849) and homogenized by vortexing. The cells were lysed by performing two passes on an M-110EH-30 microfluidizer (Microfluidics) at 7000 psi, clarified at 100K x g for 30 minutes at 4°C using an optima L-90K ultracentrifuge (Beckman), filtered (0.45 micron) and applied to a f20 mL IMAC HP column (GE Scientific) that was pre-equilibrated with lysis buffer containing 50 mM imidazole on a Bio-Rad NGC. Column was washed with lysis buffer containing 50 mM imidazole and proteins were eluted with lysis buffer containing 500 mM imidazole. After SDS-PAGE/Coomassie staining, positive fractions were pooled, dialyzed to 20 mM HEPES, 50 mM NaCl, 1 mM TCEP, 0.5 mM PMSF, 1:1000 v/v of PI, 50% glycerol, pH 7.2. Protein concentration was determined (0.88 mg/ml) and stored at −80°C for future use.
siRNA Transfection
Silencing of TDP1, TDP2 and TRIM41 were done using ON-TARGET plus SMARTpool siRNA targeting TDP1 (CAT#: L-016112-00-0005, Dharmacon), ON-TARGETplusSMARTpool siRNA targeting TDP2 (CAT#: L-017578-00-0005, Dharmacon), ON-TARGETplus SMARTpool siRNA targeting TRIM41 (CAT#: L-007105-00-0005, Dharmacon) and TRIM41 Human siRNA Oligo Duplex ((Locus ID 90933; Cat# SR314050, ORIGENE) respectively. All siRNAs were used at a final concentration of 25 nM and transfected using Lipofectamine® RNAiMAX transfection reagent (CAT#: 13778150, ThermoFisher Scientific) following the manufacturer’s protocol for 48–72 h.
Site-Directed Mutagenesis (SDM) in Mammalian Expression Vectors
Site-Directed Mutagenesis was performed using QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) following the manufacturer’s protocol. P337V TOP3B-Myc-FLAG was generated using oligonucleotides: 5′-GGTGGTCTCTGTCCGTACGTAGCTGATGTAGCCT-3′ and 5′-AGGCTACATCAGCTACGTACGGACAGAGACCACC-3′. R338W-TOP3B-Myc-FLAG was generated using oligonucleotides: 5′-GGTCTCTGTCCATGGGTAGCTGATGTAGCCT-3′ and 5′-AGGCTACATCAGCTACCCATGGACAGAGACC-3′. R338W-TOP3B-HA was generated using oligonucleotides: 5′-GGTCTCTGTCCATGGGTAGCTGATGTAGCCT-3′ and 5′-AGGCTACATCAGCTACCCATGGACAGAGACC-3′. Ubiquitin K6R was generated by Q5 SDM Kit using oligonucleotide 5′-ATCTTCGTGAGGACCCTGACTGG-3′. Ubiquitin K11R was generated using oligonucleotide 5′-CTGACTGGTAGGACCATCACTC-3′. Ubiquitin K27R was generated using oligonucleotide 5′-GAGAATGTCAGGGCAAAGATCC-3′. Ubiquitin K29R was generated using oligonucleotide 5′ GTCAAGGCAAGGATCCAAGAC-3′. Ubiquitin K33R was generated using oligonucleotide 5′-ATCCAAGACAGGGAAGGCATC-3′. Ubiquitin K48R was generated using oligonucleotide 5′-TTTGCTGGGAGACAGCTGGAA-3′. Ubiquitin K63R was generated using oligonucleotide 5′-AACATCCAGAGAGAGTCCACCC-3′. RING domain deletion mutant of human TRIM41 (deletion of amino acids 16-60; ΔRING-TRIM41) was generated by Q5 SDM Kit using oligonucleotides: 5′-CGGGAGGAGGAGGAGGAG-3′ and 5′-CTGAAGGGTCTGCACAGG-3′.
Western Blotting and antibodies
To prepare whole cell lysates for western blotting, cells were resuspended with RIPA buffer (150 mM NaCl, 1% NP- 40, 0.5% Sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 7.5, 1 mM DTT and protease inhibitor cocktail). After thorough mixing, samples were agitated at 4°C for 30 min, sonicated for 30 s with 50% pulse, centrifuged at 15,000 × g at 4°C for 15 min, and supernatants were collected.
Lysed samples were mixed with tris-glycine SDS sample buffer (Novex, LC2676) and loaded onto Novex tris-glycine gels (Novex). Blotted membranes were blocked with 5% non-fat dry milk in PBS with 0.1% Tween-20 (PBST). Primary antibodies were diluted in 5% milk in PBST by 1:1000 for Mouse monoclonal anti-FLAG M2 (Sigma-Aldrich, St. Louis, MO, CAT#: F1804), Mouse monoclonal anti-Ub (P4D1) antibody (Santa Cruz Biotechnology, Dallas, Texas, CAT#: sc-8017), 1:10000 for Rabbit anti-GAPDH monoclonal antibody (Cell Signaling Technology, Danvers, MA, CAT#: 2118S), 1: 500 for Rabbit polyclonal anti-TDP1 (Abcam, Cambridge, MA, CAT#: ab4166,), 1: 500 for Rabbit polyclonal anti-TDP2 (Bethyl, Montgomery, TX, CAT#: A302-737A), 1:500 for Rabbit polyclonal anti-TRIM41 (Abcam, Cambridge, MA, CAT#: ab98170), 1:1000 for Rabbit monoclonal anti-HA (Cell Signaling Technology, Danvers, MA, CAT#: 3724S), 1:2000 for Mouse monoclonal anti-phospho (S139)-H2AX. Secondary antibodies were diluted (1:10000) in 5% non-fat milk in PBST and signal was detected by ECL chemiluminescence reaction (Thermo Scientific, Waltham, MA).
ICE bioassay
TOP3B-DNA and -RNA cleavage complexes were isolated using the in vivo complex of enzyme (ICE) bioassay (Pourquier et al., 2000; Shaw et al., 1975). Briefly, FLAG-tagged R338W-TOP3B transfected cells were pelleted and immediately lysed with 1 mL of 1% sarkosyl. After homogenization with a Dounce, cell lysates were gently layered on step gradients containing four different CsCl (Sigma-Aldrich, CAT#:746487-1KG) solutions (2 mL of each) of the following densities: 1.82, 1.72, 1.50, and 1.45 (Shaw et al., 1975). The gradients were prepared by diltuting a stock solution of CsCl of density 1.88. Cesium sulfate (Sigma-Aldrich, CAT#:C5205-50G) was included in the bottom solution of density 1.82 to help in flotation of the RNA and sodium thiocyanate (Sigma-Aldrich, CAT#:S7757-1KG) was included in topmost solution of density 1.45 to facilitate the complete removal of non-covalently bound proteins from the sedimenting nucleic acid species (Shaw et al., 1975). Tubes were centrifuged at 30,700 rpm in a Beckman SW40 rotor for 24 h at 20°C. Half-milliliter fractions were collected from the bottom of the tubes. Fractions containing DNA and RNA were pooled separately, quantitated, diluted with 25 mM sodium phosphate buffer (pH 6.5), and applied to Immobilon-FL PVDF 0.45 μm membranes (Merck Millipore, USA, CAT#: IPFL00010) through a slot-blot vacuum manifold as described (Pourquier et al., 2000). TOP3Bccs were detected with the Mouse monoclonal anti-FLAG M2 antibody (Millipore Sigma, St. Louis, MO, CAT#: F1804).
RADAR Assay
FLAG-tagged R338W-TOP3B transfected cells (1 × 106) were washed with PBS and lysed by adding 1 mL DNAzol (ThermoFisher Scientific, CAT#:10503027). Nucleic acids were precipitated following addition of 0.5 mL of 100% ethanol, incubation at −20°C for 5 min and centrifugation (12,000 × g for 10 min). Precipitates were washed twice in 75% ethanol, resuspended in 200 μL TE buffer, heated at 65°C for 15 minutes, followed by shearing with sonication (40% power for 15 s pulse and 30 s rest 5 times). Samples were centrifuged at 15,000 rpm for 5 min and the supernatant containing nucleic acids with covalently bound proteins were collected. Nucleic acid containing protein adducts were quantitated, slot-blotted and TOP3Bccs were detected with Mouse monoclonal anti-FLAG M2 antibody (Millipore Sigma, St. Louis, MO, CAT#: F1804).
Immunoprecipitation and enrichment of RADAR assay samples
RADAR assay sample containing nucleic acids and covalent protein-nucleic acid adducts were recovered from non-transfected and FLAG-tagged WT-TOP3B transfected cells (1 × 107). Aliquots of RADAR assay samples were saved as INPUT. The rest of the RADAR samples were diluted in IP buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.2% Triton X-100, 5% glycerol, 1 mM DTT, 20 mM N-ethylmaleimide and protease inhibitor cocktail) containing anti-TOP3B antibody (abcam, Cat# ab183520) and rotated overnight at 4°C. Next day, Pierce ChIP-grade Protein A/G Magnetic Beads was added and incubated with the samples for another 4 hr. Immunoprecipitated samples were washed with IP buffer twice, resuspended in TE buffer containing 1% SDS. Nucleic acids containing TOP3Bccs were precipitated following addition of 100% ethanol and centrifugation (12,000 × g for 10 min). Precipitates were washed twice in 75% ethanol, resuspended in TE buffer. Equal volumes of resuspended nucleic acid containing protein adducts were slot-blotted and TOP3Bccs were detected with anti-TOP3B antibody (Abcam, CAT#: ab183520) (Herrero-Ruiz et al., 2020).
Detection of Ubiquitinated TOP3Bccs
For detection of ubiquitinated TOP3Bccs (Sun et al., 2019, 2020a), nucleic acids and covalent protein-nucleic acid adducts were recovered from FLAG-tagged R338W-TOP3B transfected cells using the RADAR assay. 8 μg of each RADAR assay sample was digested with 250 units micrococcal nuclease (Thermo Fisher Scientific, 100 units/μl) in the presence of 5 mM CaCl2, followed by SDS-PAGE electrophoresis for immunodetection of total TOP3Bccs and ubiquitinated TOP3Bccs. In addition, each RADAR sample was subjected to slot-blotting and immunodetection with anti-dsDNA antibody (Abcam, ab27156) to confirm equal DNA loading.
Isolation of Cellular Covalent RNA-protein Adducts from Cells Using TRIzol® Reagent
RNA-protein adducts were isolated from FLAG-tagged R338W-TOP3B transfected cells as described (Trendel et al., 2018). Briefly, 1 × 107 cells were lysed in 1 mL TRIzol Reagent (Invitrogen, USA, CAT#:15596026) by pipetting the samples up and down several times followed by incubation at room temperature for 5 min. 200 μL chloroform was added to the samples and mixed properly by inverting the tubes. After incubation at room temperature for 3 min and centrifugation for 10 minutes at 7,000 × g at 4°C, the aqueous phase was removed, and the interphase was transferred to a new tube. The interphase was gently washed twice with 1 mL low SDS buffer (Tris-Cl 50 mM, EDTA 1 mM, SDS 0.1%), resuspended in low SDS buffer, centrifuged at 5000 × g for 2 min at room temperature and the supernatant was stored. Pellets were dissolved again with 1 mL of low SDS buffer, then twice with 1 mL high SDS buffer (Tris-Cl 50 mM, EDTA 1 mM, SDS 0.5%) and all the supernatants were stored following centrifugation. NaCl was added to a final concentration of 300 mM to each of the interphase eluates, along with 10 ug of RNase-free glycogen and 1 mL isopropanol before mixing by inversion. Samples were spun down for 15 min with 18,000 × g at −10°C. Supernatant were discarded, pellet was resuspended in 70% ethanol. Samples were again centrifuged for 1 min at 18000 × g at room temperature. Supernatant were discarded, residual ethanol removed, and the pellets were resuspended in nuclease-free water at 4°C. 10X TURBO DNase Buffer (ThermoFisher Scientific, USA) was added to the resuspended samples to 1X concentration along with 10 μL TURBO DNase (ThermoFisher Scientific, USA) and incubated for 60 minutes at 37°C and 700 rpm shaking. After DNase treatment, samples were isopropanol precipitated in the presence of 300 mM NaCl and dissolved in DEPC treated water. RNA purity and concentrations were estimated by spectroscopy on a NanoDrop 1000 Spectrophotometer (ThermoFisher Scientific, USA). Samples were slot-blotted and TOP3Bccs on RNA were detected using Mouse monoclonal anti-FLAG M2 antibody (Millipore Sigma, St. Louis, MO, CAT#: F1804).
Generation of TOP3Bccs in vitro and digestion by TDP1, TDP2 and Benzonase
The hairpin substrate oligo nucleotide with long 3′-tail: GGGATTATTGAACTGTTGTTCAAACTTTAGAACTAGCCATCCGATTT ACACTTTGCCCCTATCCACCCC was synthesized by IDT. 300 nM of annealed substrate was combined with 4 uM of purified recombinant TOP3B in 100 mM potassium glutamate (pH 7.0), 3 mM MgCl2, 0.02% v/v Tween-20, 1 mM DTT, and incubated at 30°C for 15 mins before addition of 1 or 3 μM of TDP1 or TDP2 and incubated for an additional 60 min at 25°C. Benzonase (3 or 9 Units) was used as positive control. SDS (0.2%) was added to the samples to stop the reaction. The samples were resolved on 6% tris-glycine-SDS-PAGE and western blotting was carried out using standard techniques with rabbit monoclonal anti-TOP3B (Abcam, CAT#: ab183520).
TOP3B mediated cleavage of modified D3+ extended oligonucleotide Substrate
Oligonucleotide substrates were labeled on the 5′ end with [γ-32P] ATP and T4 Polynucleotide Kinase before passing through mini Quick Spin Oligo Columns (Roche) and annealed after heating at 95°C for 5 minutes. 10 nM of labeled substrate was incubated with 22 nM of recombinant TOP3B in 100 mM potassium glutamate (pH 7.0), 3 mM MgCl2, 0.02% v/v Tween-20 and 1 mM DTT, incubated at 37°C for 30 minutes before addition of 0.2% SDS and 1 volume of gel loading buffer [96% (v/v) formamide, 10 mM ethylenedia-minetetraacetic acid, 1% (w/v) xylene cyanol and 1% (w/v) bromophenol blue]. Samples were analyzed by 20% denaturing polyacrylamide gel electrophoresis gels, which were dried and exposed on PhosphorImager screens. Imaging was done using a Typhoon 8600 and ImageQuant software (GE Healthcare, UK).
Denaturation of TOP3Bcc and processing by TDP2
300 nM of D3+ extended substrate with biotin label on the 3′ end was incubated with 4 μM of TOP3γ in 100 mM potassium glutamate (pH 7.0), 3 mM MgCl2, 0.02% v/v Tween-20, 1 mM DTT at 30°C for 30 minutes before addition of 0.2% SDS. The samples were ethanol precipitated following standard protocols and resuspended in 8 mM NaOH. The recovered mixture was incubated with TDP2 (0, 2.25, 4.5 and 9 μM) in 100 mM potassium glutamate (pH 7.0), 3 mM MgCl2, 0.02% v/v Tween-20 and 1 mM DTT at 25°C overnight. The samples were then combined with 1 volume of Tris-Glycine-SDS sample buffer supplemented with 4% 2-mercaptoethanol and resolved on SDS-PAGE (6%). The resolved samples were transferred onto PVDF membrane and probed with anti-TOP3B antibody and IR Dye-coupled streptavidin following standard procedures. The images were detected by a Bio-Rad ChemiDoc Imager.
Proteinase K-digestion of TOP3Bcc and processing by TDP2
D3+ extended oligonucleotide substrates were labeled on the 3′ end with [α-32P] cordycepin 5′-triphosphate and terminal deoxynucleotidyl transferase before passing through mini Quick Spin Oligo Columns (Roche) and annealed after heating at 95°C for 5 minutes. The labeled oligonucleotide substrates (80 nM) were incubated with TOP3B (4 μM) in 100 mM potassium glutamate (pH 7.0), 3 mM MgCl2, 0.02% v/v Tween-20 and 1 mM DTT at 30°C for 30 minutes. Proteinase K (0.6 mg/mL) was added to the samples and incubated at 30°C overnight before the heat inactivation of Proteinase K at 95°C for 5 minutes. TDP2 (1 μM) was added the samples and incubated at 25°C for 30 minutes. 1 volume of gel loading buffer was added to samples [96% (v/v) formamide, 10 mM ethylenedia-minetetraacetic acid, 1% (w/v) xylene cyanol and 1% (w/v) bromophenol blue] before analysis on a 20% denaturing polyacrylamide gel electrophoresis gels, which were dried and exposed on PhosphorImager screens. Imaging was done using a Typhoon 8600 and ImageQuant software (GE Healthcare, UK).
Immunoprecipitation of RADAR assay samples and detection of ubiquitinated TOP3B cleavage complexes
RADAR assay sample containing nucleic acids and covalent protein-nucleic acid adducts were recovered from non-transfected, FLAG-tagged WT-TOP3B and R338W-TOP3B transfected cells (1 × 106). Aliquots (16 μg) of RADAR assay samples were saved as INPUT. The rest of the RADAR samples were diluted in IP buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.2% Triton X-100, 5% glycerol, 1 mM DTT, 20 mM N-ethylmaleimide and protease inhibitor cocktail) containing anti-TOP3B antibody (abcam, Cat# ab183520) and rotated overnight at 4°C. Next day, Pierce ChIP-grade Protein A/G Magnetic Beads was added and incubated with the samples for another 4 hr. Immunoprecipitated samples were washed with IP buffer twice, resuspended in TE buffer containing 5 mM CaCl2 and excess amount of micrococcal nuclease (Thermo Fisher Scientific, 100 units/μl). Samples were incubated in Thermomixer (1200 rpm, 37°C for 4 hr), resuspended in tris-glycine SDS sample buffer (Novex, LC2676) for SDS-PAGE and immunoblotted with different antibodies as indicated for detection of total TOP3Bccs and ubiquitinated TOP3Bccs. 8 μg of each INPUT RADAR assay sample were also digested with 250 units micrococcal nuclease (Thermo Fisher Scientific, 100 units/μl) in the presence of 5 mM CaCl2, followed by SDS-PAGE electrophoresis for immunodetection of total TOP3Bccs and ubiquitinated TOP3Bccs. In addition, each INPUT RADAR sample was subjected to slot-blotting and immunodetection with anti-dsDNA antibody (Abcam, ab27156) to confirm equal DNA loading.
R-loop detection by DOT-BLOT method using s9.6 Ab
For R-loop detection by slot-blot, genomic DNA was extracted from non-transfected, FLAG-tagged WT-TOP3B and R338W-TOP3B transfected HCT116 and HEK293 cells as described previously (Castillo-Guzman et al., 2020; Halász et al., 2017; Manzo et al., 2018; Sanz and Chédin, 2019). Briefly, cells were lysed in TE buffer containing 0.6% SDS and proteinase K (at 37°C overnight), phase separated using phenol/chloroform/isoamyl alcohol (25:24:1), ethanol precipitated and resuspended in TE buffer. Genomic DNA was sheared by sonication (40% output for 20 s pulse and 60 s rest for 10 times) and again purified by phenol/chloroform/isoamyl alcohol (25:24:1) extraction. Genomic DNA (8 μg) were spotted on a nitrocellulose membrane, crosslinked with UV light (120 mJ/cm2)), blocked with PBS-Tween (0.1%) buffer and 5% non-fat milk (Room temperature for 1hr) and incubated with mouse S9.6 antibody (1:500 dilution, overnight at 4°C, Millipore Sigma, Cat# MABE1095). After washing with PBS-Tween (0.1%), membrane was incubated with HRP-conjugated anti-mouse secondary antibody, washed and developed with ECL techniques. In case of RNase H treated control, 8 genomic DNA was pre-incubated with 16 U of RNase H for three hours at 37°C.
Colony Formation Assay
HCT116 and HEK293 cells were transfected with R338W-TOP3B, WT-TOP3B or mock-transfection reagent (NT) for three days and after harvesting cells were plated in 6-well plates (500 cells/ well) in triplicate. After 14 days, colonies were fixed, stained with crystal violet, and well intensity was measured using the Fiji software. Data were normalized to the mock-transfection (NT) conditions.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantifications were carried out using the Fiji software. Data are provided as means ± standard deviations (SD) from the number of independent experiments performed. Statistical analyses and graphical representation were carried out using GraphPad prism 7 software. Statistical test methods are described in each figure legend (Figures 7F and 7H). Statistical significance is represented by (***) and indicates a computed p value < 0.0005 (Figures 7F and 7H).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal ANTI-FLAG® M2 antibody, Sigma-Aldrich | Sigma-Aldrich | Cat# F1804, RRID: AB_262044 |
| Ub (P4D1) antibody, Santa Cruz Biotechnology | Santa Cruz Biotechnology | Cat# sc-8017, RRID:AB_628423 |
| Rabbit Anti-GAPDH Monoclonal Antibody, Unconjugated, Clone 14C10 | Cell Signaling Technology | Cat# 2118, RRID:AB_561053 |
| Rabbit Anti-TDP1 Polyclonal Antibody, Unconjugated, Abcam | Abcam | Cat# ab4166, RRID:AB_304337 |
| TDP2 Antibody, Bethyl | Bethyl | Cat# A302-737A, RRID:AB_10631698 |
| TRIM41 antibody, Abcam | Abcam | Cat# ab98170, RRID:AB_10672571 |
| Anti-TOP3B antibody [EP7779] - C-terminal (ab183520) | Abcam | Cat# ab183520 |
| Rabbit Anti-HA-Tag Monoclonal Antibody, Unconjugated, Clone C29F4 | Cell Signaling Technology | Cat# 3724, RRID:AB_1549585 |
| Sheep Anti-Mouse IgG ECL Antibody, HRP Conjugated, GE Healthcare | GE Healthcare | Cat# NA9310-1ml, RRID:AB_772193 |
| Donkey Anti-Rabbit IgG ECL Antibody, HRP Conjugated, GE Healthcare | GE Healthcare | Cat# NA9340-1ml, RRID:AB_772191 |
| Anti-ds DNA antibody | Abcam | Cat# ab27156, RRID:AB_470907 |
| Anti-DNA-RNA Hybrid Antibody, clone S9.6 | Millipore Sigma | Cat# MABE1095, |
| Mouse monoclonal anti-phospho (S139)-H2AX (JBW301) | Millipore Sigma | Cat# 05-636, RRID: AB_309864 |
| Bacterial and Virus Strains | ||
| NEB® 5-alpha Competent E. coli (High Efficiency) | NEW ENGLAND BioLabs Inc. | Cat# C2987H |
| MAX Efficiency DH10B Competent Cells | ThermoFisher Scientific | Cat# 18297010 |
| DE77, a DH10Bac-derived strain | Bac-to-Bac system, Thermo Fisher | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM - Dulbecco’s Modified Eagle Medium | ThermoFisher Scientific | Cat# 11965-092 |
| Fetal Bovine Serum | Gemini | Cat# 100-106 |
| Penicillin-Streptomycin | ThermoFisher Scientific | Cat# 15140-122 |
| Trypsin-EDTA (0.05%) | ThermoFisher Scientific | Cat# 25300054 |
| cOmplete Mini, EDTA-free (protease inhibitor cocktail) | Roche | Cat# 11836170001 |
| Recombinant Human TOP3B | This Paper | N/A |
| InSolution MG-132 - CAS 133407-82-6 - Calbiochem | Sigma-Aldrich | Cat# 474791-5MG |
| TAK243 (MLN7243) | Selleckchem | Cat# S8341 |
| PEI | Polysciences | Cat# 23966 |
| Lipofectamine 3000 Reagent | ThermoFisher Scientific | Cat# L3000015 |
| Lipofectamine® RNAiMAX transfection reagent | ThermoFisher Scientific | Cat# 13778150 |
| Benzonase | Sigma-Aldrich | Cat# E8263 |
| Pierce ChIP-grade Protein A/G Magnetic Beads | ThermoFisher Scientific | Cat# 26162 |
| tris-glycine SDS sample buffer | Novex | Cat# LC2676 |
| SuperSignal West Femto Maximum Sensitivity Substrate | ThermoFisher Scientific | Cat# 34095 |
| TRIzol Reagent | ThermoFisher Scientific | Cat# 15596026 |
| Micrococcal Nuclease Solution (≥100 U/μL) | ThermoFisher Scientific | Cat# 88216 |
| RNase A | ThermoFisher Scientific | Cat# EN0531 |
| RNase T1 | ThermoFisher Scientific | Cat# EN0542 |
| Invitrogen TURBO DNase (2 U/μL) | ThermoFisher Scientific | Cat# AM2238 |
| N-Ethylmaleimide | Millipore Sigma | Cat# E3876-25G |
| Cesium chloride (CsCl) | Sigma-Aldrich | Cat# 746487-1KG |
| Cesium sulfate | Sigma-Aldrich | Cat# C5205-50G |
| Sodium thiocyanate | Sigma-Aldrich | Cat# S7757-1KG |
| DNAzol | ThermoFisher Scientific | Cat# 10503027 |
| Glycogen, RNA grade | ThermoFisher Scientific | Cat# R0551 |
| Invitrogen, Proteinase K Solution | ThermoFisher Scientific | Cat# 4333793 |
| Invitrogen, Proteinase K Solution (20 mg/mL), RNA grade | ThermoFisher Scientific | Cat# 25530049 |
| N-Lauroylsarcosine sodium salt | Millipore Sigma | Cat# L9150 |
| Critical Commercial Assays | ||
| QuikChange II XL site-directed mutagenesis kit | Agilent Technologies | Cat# 200521 |
| Q5® Site-Directed Mutagenesis Kit | NEW ENGLAND BioLabs Inc. | Cat# E0554S |
| Deposited data | ||
| Raw imaging data | This paper; Mendeley Data | https://doi.org/10.17632/jp9vs3sddy.1 |
| Experimental Models: Cell Lines | ||
| HEK293 | ATCC | CRL-1573 |
| HCT116 | Developmental Therapeutics Program (NCI/NIH) | N/A |
| Oligonucleotides | ||
| Hairpin substrate oligonucleotide with long 3′-tail (Modified D3+ extended oligo): 5′-GGGATT ATTGAACTGTTGTTCAAACTTTAGAACTAGCCA TCCGATTTACACTTTGCCCCTATCCACCCC-3′ |
IDT oligo | N/A |
| D3+ oligo (DiGate and Marians, 1992): 5′-GGGATTATTGAACTGTTGTTCA AGCGTGGT-3′ |
IDT oligo | N/A |
| siRNA targeting sequences, Human TDP1, SMART pool GGAGUUAAGCCAAAGUAUAUCAGUU ACUUGAUGGCUUAGACCAUAUCUAGUAGU GAUCUAGACAGUUUCAAAGUGA |
Dharmacon | Cat# L-016112-00-0005 |
| siRNA targeting sequences, Human TDP2, SMART pool GUACAGCCCAGAUGUGAUAGCAGAA GAGGGACACAUUAUCUAAGGGAUCGAGA GGUUAAAGGGCUCUGAACUCCUA |
Dharmacon | Cat# L-017578-00-0005 |
| siRNA targeting sequences, Human TRIM41, SMART pool (A mixture of 4 siRNA) TRIM41 siRNA 1: CAAUAGGUG UGAAGAGGUATRIM41 siRNA 2: CCAAUAUGGUCCAGGUGAUTRIM41 siRNA 3: GAGAUGAGUUAGAUCGGGATRIM41 siRNA 4: UAGCUUCACUUGAGAGAGA |
Dharmacon, Horizon Discovery | Cat# L-007105-00-0005 |
| TRIM41 Human siRNA Oligo Duplex (Locus ID 90933): TRIM41 (Human) - 3 unique 27-mer siRNA duplexes SR314050A: rGrArArGrCrUrCrUrUrCrUr GrCrGrArGrGrUrArGrArCrGAA SR314050B: rArGrArGrCrCrUrUrGrGrGrUrArUrArArUrCrUrAr UrUrUTT SR314050C: rGrUrUrGrArGrGrArArGr ArGrGrArUrCrUrArGrArCrCrCCG |
ORIGENE | Cat# SR314050 |
| Primers for cloning and mutagenesis: See Table S1 | N/A | N/A |
| Recombinant DNA | ||
| Human TOP3B-Myc-FLAG | OriGene | Cat# RC223204 |
| Human TRIM41-Myc-FLAG | OriGene | Cat# RC210557 |
| human pCMV6-AN-DDK(Flag)-hTDP2 ORF clone | Huang et al., 2018b | N/A |
| pcDNA3-TOP3B-HA | This paper | N/A |
| pcDNA3-TDP2-HA | This paper | N/A |
| pRK5-HA-Ubiquitin-WT | Addgene (gift from Ted Dawson) | Cat# 17608 |
| pENTR3C-TOP3B | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism 7 (software for drawing graphs and statistics analysis) | GraphPad | N/A |
| Fiji | Schindelin et al., 2012 | https://fiji.sc |
Highlights.
Generation of R338W-TOP3B mutant for in vivo detection of TOP3Bccs in DNA and RNA
Cellular TOP3Bccs result in growth defect, R-loop accumulation, and genomic damage
TRIM41 acts as E3 ubiquitin ligase for TOP3Bcc prior to proteasomal processing
TDP2 processes both RNA and DNA TOP3Bccs
ACKNOWLEDGMENTS
We thank Protein Expression Laboratory (Protein and Nucleic Acid Production–Center for Cancer Research [CCR]), NCI-Frederick, Maryland, for helping in the production of recombinant human TOP3B. Our studies are supported by the Center for Cancer Research, the Intramural Program of the National Cancer Institute, NIH, Bethesda, MD 20892 (Z01 BC 006161-17 and Z01 BC 006150-19).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108569.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ahmad M, Xue Y, Lee SK, Martindale JL, Shen W, Li W, Zou S, Ciaramella M, Debat H, Nadal M, et al. (2016). RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals. Nucleic Acids Res. 44, 6335–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Shen W, Li W, Xue Y, Zou S, Xu D, and Wang W (2017a). Topoisomerase 3β is the major topoisomerase for mRNAs and linked to neuro-development and mental dysfunction. Nucleic Acids Res. 45, 2704–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Xu D, and Wang W (2017b). Type IA topoisomerases can be “magicians” for both DNA and RNA in all domains of life. RNA Biol. 14, 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L, Lowe J, Dick LR, Mayer RJ, and Brownell JE (2011). Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov 10, 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Guzman D, Hartono SR, Sanz LA, and Chédin F (2020). SF3B1-targeted Splicing Inhibition Triggers Global Alterations in Transcriptional Dynamics and R-Loop Metabolism. bioRxiv, 2020.2006.2008.130583. [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, and Varshavsky A (1989). A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583. [DOI] [PubMed] [Google Scholar]
- Chen D, Gould C, Garza R, Gao T, Hampton RY, and Newton AC (2007). Amplitude control of protein kinase C by RINCK, a novel E3 ubiquitin ligase. J. Biol. Chem 282, 33776–33787. [DOI] [PubMed] [Google Scholar]
- Cheng B, Shukla S, Vasunilashorn S, Mukhopadhyay S, and Tse-Dinh YC (2005). Bacterial cell killing mediated by topoisomerase I DNA cleavage activity. J. Biol. Chem 280, 38489–38495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Annamalai T, Sorokin E, Abrenica M, Aedo S, and Tse-Dinh YC (2009). Asp-to-Asn substitution at the first position of the DxD TOPRIM motif of recombinant bacterial topoisomerase I is extremely lethal to E. coli. J. Mol. Biol 385, 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SC, Meagher M, Kassouf N, Hafezparast M, McKinnon PJ, Haywood R, and El-Khamisy SF (2017). Mitochondrial protein-linked DNA breaks perturb mitochondrial gene transcription and trigger free radical-induced DNA damage. Sci. Adv 3, e1602506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, and Caldecott KW (2009). A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 461, 674–678. [DOI] [PubMed] [Google Scholar]
- Daghsni M, Lahbib S, Fradj M, Sayeb M, Kelmemi W, Kraoua L, Kchaou M, Maazoul F, Echebbi S, Ben Ali N, et al. (2018). TOP3B: A Novel Candidate Gene in Juvenile Myoclonic Epilepsy? Cytogenet. Genome Res 154, 1–5. [DOI] [PubMed] [Google Scholar]
- DiGate RJ, and Marians KJ (1992). Escherichia coli topoisomerase III-catalyzed cleavage of RNA. J. Biol. Chem 267, 20532–20535. [PubMed] [Google Scholar]
- Gao R, Huang SY, Marchand C, and Pommier Y (2012). Biochemical characterization of human tyrosyl-DNA phosphodiesterase 2 (TDP2/TTRAP): a Mg(2+)/Mn(2+)-dependent phosphodiesterase specific for the repair of topoisomerase cleavage complexes. J. Biol. Chem 287, 30842–30852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Schellenberg MJ, Huang SY, Abdelmalak M, Marchand C, Nitiss KC, Nitiss JL, Williams RS, and Pommier Y (2014). Proteolytic degradation of topoisomerase II (Top2) enables the processing of Top2·DNA and Top2·RNA covalent complexes by tyrosyl-DNA-phosphodiesterase 2 (TDP2). J. Biol. Chem 289, 17960–17969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Bhattacharjee S, Chowdhuri SP, Mallick A, Rehman I, Basu S, and Das BB (2019). SCAN1-TDP1 trapping on mitochondrial DNA promotes mitochondrial dysfunction and mitophagy. Sci. Adv 5, eaax9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto-Ito S, Yamagata A, Takahashi TS, Sato Y, and Fukai S (2017). Structural basis of the interaction between Topoisomerase IIIβ and the TDRD3 auxiliary factor. Sci. Rep 7, 42123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halász L, Karányi Z, Boros-Oláh B, Kuik-Rózsa T, Sipos É, Nagy É, Mosolygó-L Á, Mázló A, Rajnavölgyi É, Halmos G, and Székvölgyi L (2017). RNA-DNA hybrid (R-loop) immunoprecipitation mapping: an analytical workflow to evaluate inherent biases. Genome Res. 27, 1063–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S (2017). TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem. Sci 42, 297–311. [DOI] [PubMed] [Google Scholar]
- Herrero-Ruiz A, Martínez-García P, Terrón-Bautista J, Lieberman JA, Jimeno-González S, and Cortés-Ledesma F (2020). Control of RNA polymerase II promoter-proximal pausing by DNA supercoiling. bioRxiv, 2020.2005.2012.091058. [Google Scholar]
- Hoa NN, Shimizu T, Zhou ZW, Wang ZQ, Deshpande RA, Paull TT, Akter S, Tsuda M, Furuta R, Tsutsui K, et al. (2016). Mre11 Is Essential for the Removal of Lethal Topoisomerase 2 Covalent Cleavage Complexes. Mol. Cell 64, 580–592. [DOI] [PubMed] [Google Scholar]
- Huang L, Wang Z, Narayanan N, and Yang Y (2018a). Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic Acids Res. 46, 3061–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SN, Dalla Rosa I, Michaels SA, Tulumello DV, Agama K, Khiati S, Jean SR, Baechler SA, Factor VM, Varma S, et al. (2018b). Mitochondrial tyrosyl-DNA phosphodiesterase 2 and its TDP2S short isoform. EMBO Rep. 19, e42139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyer ML, Milhollen MA, Ciavarri J, Fleming P, Traore T, Sappal D, Huck J, Shi J, Gavin J, Brownell J, et al. (2018). A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med 24, 186–193. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, and Varshavsky A (1995). A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem 270, 17442–17456. [DOI] [PubMed] [Google Scholar]
- Katyal S, Lee Y, Nitiss KC, Downing SM, Li Y, Shimada M, Zhao J, Russell HR, Petrini JH, Nitiss JL, and McKinnon PJ (2014). Aberrant topoisomerase-1 DNA lesions are pathogenic in neurodegenerative genome instability syndromes. Nat. Neurosci 17, 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman CS, Genovese A, and Butler MG (2016). Deletion of TOP3B Is Associated with Cognitive Impairment and Facial Dysmorphism. Cytogenet. Genome Res 150, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawale AS, and Povirk LF (2018). Tyrosyl-DNA phosphodiesterases: rescuing the genome from the risks of relaxation. Nucleic Acids Res. 46, 520–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiianitsa K, and Maizels N (2013). A rapid and sensitive assay for DNA-protein covalent complexes in living cells. Nucleic Acids Res. 41, e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, and Hanai R (2001). M phase-specific association of human topoisomerase IIIbeta with chromosomes. Biochem. Biophys. Res. Commun 287, 282–287. [DOI] [PubMed] [Google Scholar]
- Kravtsova-Ivantsiv Y, and Ciechanover A (2012). Non-canonical ubiquitin-based signals for proteasomal degradation. J. Cell Sci 125, 539–548. [DOI] [PubMed] [Google Scholar]
- Kwan KY, and Wang JC (2001). Mice lacking DNA topoisomerase IIIbeta develop to maturity but show a reduced mean lifespan. Proc. Natl. Acad. Sci. USA 98, 5717–5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Moens PB, and Wang JC (2003). Infertility and aneuploidy in mice lacking a type IA DNA topoisomerase III beta. Proc. Natl. Acad. Sci. USA 100, 2526–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Greenwald RJ, Mohanty S, Sharpe AH, Shaw AC, and Wang JC (2007). Development of autoimmunity in mice lacking DNA topoisomerase 3beta. Proc. Natl. Acad. Sci. USA 104, 9242–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassot I, Mora S, Lesage S, Zieba BA, Coque E, Condroyer C, Bossowski JP, Mojsa B, Marelli C, Soulet C, et al. (2018). The E3 Ubiquitin Ligases TRIM17 and TRIM41 Modulate alpha-Synuclein Expression by Regulating ZSCAN21. Cell Rep. 25, 2484–2496.e2489. [DOI] [PubMed] [Google Scholar]
- Lee SK, Xue Y, Shen W, Zhang Y, Joo Y, Ahmad M, Chinen M, Ding Y, Ku WL, De S, et al. (2018). Topoisomerase 3β interacts with RNAi machinery to promote heterochromatin formation and transcriptional silencing in Drosophila. Nat. Commun 9, 4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzo SG, Hartono SR, Sanz LA, Marinello J, De Biasi S, Cossarizza A, Capranico G, and Chedin F (2018). DNA Topoisomerase I differentially modulates R-loops across the human genome. Genome Biol. 19, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell A (1999). DNA gyrase as a drug target. Biochem. Soc. Trans 27, 48–53. [DOI] [PubMed] [Google Scholar]
- Meroni G, and Diez-Roux G (2005). TRIM/RBCC, a novel class of ’single protein RING finger’ E3 ubiquitin ligases. BioEssays 27, 1147–1157. [DOI] [PubMed] [Google Scholar]
- Meyer HJ, and Rape M (2014). Enhanced protein degradation by branched ubiquitin chains. Cell 157, 910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano LM, and Meroni G (2012). TRIM family: Pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life 64, 64–71. [DOI] [PubMed] [Google Scholar]
- Narula G, Annamalai T, Aedo S, Cheng B, Sorokin E, Wong A, and Tse-Dinh YC (2011). The strictly conserved Arg-321 residue in the active site of Escherichia coli topoisomerase I plays a critical role in DNA rejoining. J. Biol. Chem 286, 18673–18680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL (2009). Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Costa JP, Zanetti J, Oliveira LR, Soares FA, Ramalho LZ, Silva Ramalho F, Garcia SB, and Ribeiro-Silva A (2010). Significance of topoisomerase IIIβ expression in breast ductal carcinomas: strong associations with disease-specific survival and metastasis. Hum. Pathol 41, 1624–1630. [DOI] [PubMed] [Google Scholar]
- Patil G, Xu L, Wu Y, Song K, Hao W, Hua F, Wang L, and Li S (2020). TRIM41-Mediated Ubiquitination of Nucleoprotein Limits Vesicular Stomatitis Virus Infection. Viruses 12, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Huang SY, Gao R, Das BB, Murai J, and Marchand C (2014). Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair (Amst.) 19, 114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, and Nitiss JL (2016). Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol 17, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot JJ, Yao KC, Robertson CA, and Nash HA (1999). Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286, 552–555. [DOI] [PubMed] [Google Scholar]
- Pourquier P, Takebayashi Y, Urasaki Y, Gioffre C, Kohlhagen G, and Pommier Y (2000). Induction of topoisomerase I cleavage complexes by 1-beta -D-arabinofuranosylcytosine (ara-C) in vitro and in ara-C-treated cells. Proc. Natl. Acad. Sci. USA 97, 1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. (2014). A proteome-scale map of the human interactome network. Cell 159, 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A, and Tanaka K (2009). Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 28, 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz LA, and Chédin F (2019). High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc 14, 1734–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H, Tsuda M, Morimoto S, Saha LK, Rahman MM, Kiyooka Y, Fujiike H, Cherniack AD, Itou J, Callen Moreu E, et al. (2018). BRCA1 ensures genome integrity by eliminating estrogen-induced pathological topoisomerase II-DNA complexes. Proc. Natl. Acad. Sci. USA 115, E10642–E10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg MJ, Lieberman JA, Herrero-Ruiz A, Butler LR, Williams JG, Muñoz-Cabello AM, Mueller GA, London RE, Cortés-Ledesma F, and Williams RS (2017). ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links. Science 357, 1412–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg MJ, Appel CD, Riccio AA, Butler LR, Krahn JM, Liebermann JA, Cortés-Ledesma F, and Williams RS (2020). Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2. Nucleic Acids Res. 48, 6310–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JL, Blanco J, and Mueller GC (1975). Simple procedure for isolation of DNA, RNA and protein fractions from cultured animal cells. Anal. Biochem 65, 125–131. [DOI] [PubMed] [Google Scholar]
- Siaw GE, Liu IF, Lin PY, Been MD, and Hsieh TS (2016). DNA and RNA topoisomerase activities of Top3β are promoted by mediator protein Tudor domain-containing protein 3. Proc. Natl. Acad. Sci. USA 113, E5544–E5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll G, Pietiläinen OPH, Linder B, Suvisaari J, Brosi C, Hennah W, Leppä V, Torniainen M, Ripatti S, Ala-Mello S, et al. (2013). Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci 16, 1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jenkins LMM, Su YP, Nitiss KC, Nitiss JL, and Pommier Y (2019). A conserved SUMO-Ubiquitin pathway directed by RNF4/SLX5-SLX8 and PIAS4/SIZ1 drives proteasomal degradation of topoisomerase DNA-protein crosslinks. bioRxiv. 10.1101/707661. [DOI] [Google Scholar]
- Sun Y, Miller Jenkins LM, Su YP, Nitiss KC, Nitiss JL, and Pommier Y (2020a). A conserved SUMO pathway repairs topoisomerase DNA-protein cross-links by engaging ubiquitin-mediated proteasomal degradation. Sci. Adv 6, eaba6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Saha LK, Saha S, Jo U, and Pommier Y (2020b). Debulking of topoisomerase DNA-protein crosslinks (TOP-DPC) by the proteasome, non-proteasomal and non-proteolytic pathways. DNA Repair (Amst.) 94, 102926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Saha S, Wang W, Saha LK, Huang SN, and Pommier Y (2020c). Excision repair of topoisomerase DNA-protein crosslinks (TOP-DPC). DNA Repair (Amst.) 89, 102837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Fukuda Y, Mashima K, and Hanai R (2005). Intracellular localization and domain organization of human TRIM41 proteins. Mol. Biol. Rep 32, 87–93. [DOI] [PubMed] [Google Scholar]
- Trendel J, Schwarzl T, Prakash A, Bateman A, Hentze MW, and Krijgsveld J (2018). The Human RNA-Binding Proteome and Its Dynamics During Arsenite-Induced Translational Arrest. bioRxiv. 10.1016/j.cell.2018.11.004. [DOI] [PubMed] [Google Scholar]
- Virgen-Slane R, Rozovics JM, Fitzgerald KD, Ngo T, Chou W, van der Heden van Noort GJ, Filippov DV, Gershon PD, and Semler BL (2012). An RNA virus hijacks an incognito function of a DNA repair enzyme. Proc. Natl. Acad. Sci. USA 109, 14634–14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Tretter EM, Schmidt BH, and Berger JM (2011). All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol 12, 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Sali T, and Hsieh TS (2002). Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIbeta. Proc. Natl. Acad. Sci. USA 99, 7974–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Shen W, Guo R, Xue Y, Peng W, Sima J, Yang J, Sharov A, Srikantan S, Yang J, et al. (2013). Top3β is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci 16, 1238–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, and Bedford MT (2014). Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 53, 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Wallis M, Petrovic V, Challis J, Kalitsis P, and Hudson DF (2019). Loss of TOP3B leads to increased R-loop formation and genome instability. Open Biol. 9, 190222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Imaging data supporting the current study have been deposited at Mendeley Data (https://doi.org/10.17632/jp9vs3sddy.1).