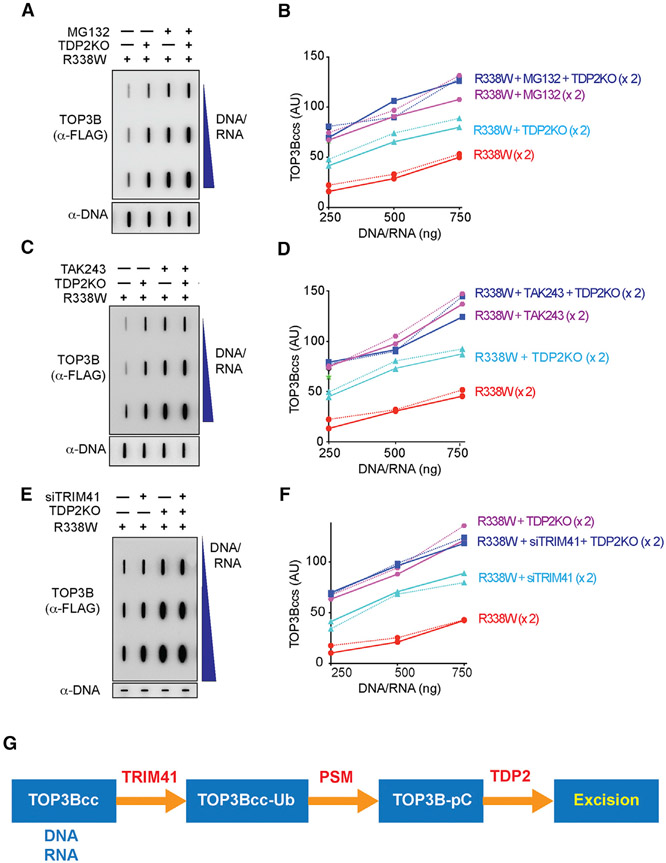
Figure 6. TDP2-Mediated Repair of TOP3Bccs Is Dependent on Ubiquitination and Proteasomal Processing.
(A) WT and TDP2KO HCT116 cells were transfected with FLAG-tagged R338W-TOP3B alone and incubated for 72 h. Before harvest, cells were treated with MG132 (10 μM, 2 h) as indicated. Nucleic acids and protein-nucleic acid adducts were recovered by RADAR assay. TOP3Bccs were detected using anti-FLAG antibody. Equal loading was with anti-dsDNA antibody. The figure is representative of two independent experiments.
(B) Quantitation of TOP3Bccs in two independent experiments as shown in (A).
(C) WT and TDP2KO HCT116 cells were transfected with FLAG-tagged R338W-TOP3B alone and incubated for 72 h. Before harvest, cells were treated with TAK243 (10 μM, 2 h). Nucleic acids and protein-nucleic acid adducts were recovered by RADAR assay, and TOP3Bccs were detected using anti-FLAG antibody. Equal loading was tested with anti-dsDNA antibody. The figure is representative of two independent experiments.
(D) Quantitation of TOP3Bccs in two independent RADAR assays as shown in (C).
(E) WT and TDP2KO HCT116 cells were transfected with FLAG-tagged R338W-TOP3B alone or co-transfected with siTRIM41 constructs and incubated for 72 h. TOP3Bccs were detected after RADAR assay with anti-FLAG antibody. Equal loading was tested with anti-dsDNA antibody. The figure is representative of two independent experiments.
(F) Quantitation of TOP3Bccs in two independent RADAR assays as shown in (E).
(G) Model for the processing of TOP3Bccs by TRIM14, the proteasome (PSM), and TDP2. See also Figure S6.