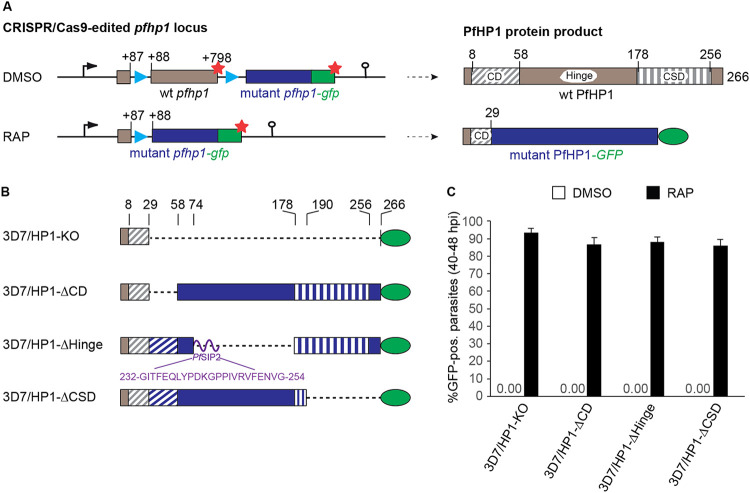
FIG 1.
Generation of DiCre-inducible PfHP1 truncation mutants. (A) Schematics of the CRISPR/Cas9-edited pfhp1 locus (left) and corresponding PfHP1 protein products (right) before (DMSO) and after (RAP) rapamycin-induced DiCre-dependent excision of the floxed wild-type (wt) pfhp1 locus. Blue arrowheads indicate the position of sera2 intron:loxP elements. Red stars indicate stop codons. Brown and blue boxes represent the wild-type and the replacing mutant sequences pfhp1/PfHP1, respectively. Green boxes represent the gfp/GFP sequence. The CD, hinge domain, and CSD within wild-type PfHP1 are indicated. Numbers in the gene and protein schematics refer to nucleotide and amino acid positions, respectively. (B) Diagrams showing the organization of the PfHP1 truncation mutants expressed in the 3D7/HP1-KO, 3D7/HP1-ΔCD, 3D7/HP1-ΔHinge, and 3D7/HP1-ΔCSD lines after RAP treatment. Dashed lines represent corresponding deletions in the mutant PfHP1 protein sequences. Brown and blue colors represent the remaining wild-type PfHP1 N terminus and the replacing mutant protein sequences, respectively. The CD and CSD are indicated by diagonal and vertical dashed stripes, respectively. The purple curved line represents the short PfSIP2-derived linker polypeptide between the CD and CSD in the PfHP1-ΔHinge mutant. The amino acid sequence of this linker and its position within the PfSIP2 protein are indicated. Numbers on top indicate amino acid positions within wild-type PfHP1. (C) Proportion of GFP-positive parasites observed 40 h after treatment with RAP or DMSO (control). Values represent the means of results from three independent biological replicates (error bars indicate SD). For each sample, >200 iRBCs were counted. pos., positive.