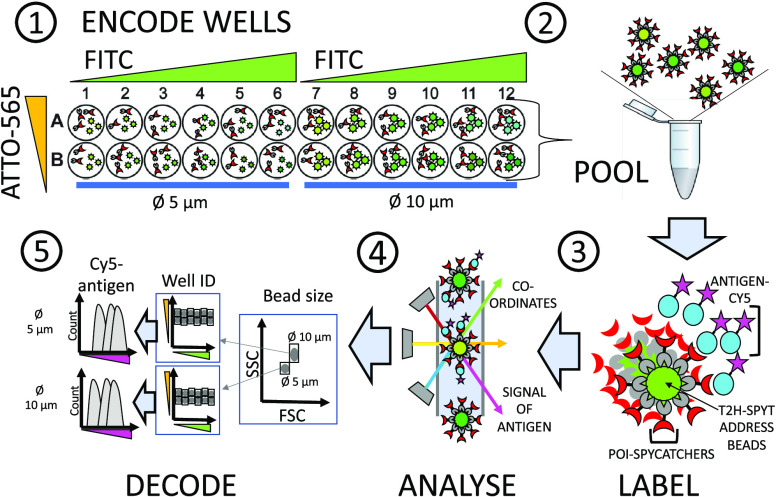
Figure 2.
Schematic workflow for screening POI-SpyCatcher clones with a 24-plex spectrally encoded SBA functionalized with SpyTag. (1) Encoding expression wells with address beads. POI-SpyCatcher fusions were expressed in E. coli cells in 96-well microtiter plates. After cell lysis and pelleting the debris, the fusion proteins in the soluble fraction were directly captured on the beads. Each well containing the expression supernatant was supplemented with 104 address beads labeled with different quantities of fluorescent dyes, FITC and bio-ATTO-565, coding for the column and row identity, respectively. Consequently, the source well of the probed bead-bound POIs could be later traced by flow cytometry. (2, 3) Pooling and labeling. After POI-SpyCatcher fusion proteins were stably captured on the bead surface by the formation of isopeptide bonds, the beads were washed to remove the unbound POI-SpyCatchers and pooled for labeling with the Cy5 antigen. (4) Flow cytometry analysis. Readout via two laser lines (blue, λ = 488 nm for detecting FITC; yellow, λ = 563 nm, for detecting ATTO-565) decoded the source well coordinates of the examined bead, and a third laser (red, λ = 633 nm) detected the signal indicative of antigen-binding (Cy5). (5) Gating and decoding. The recorded events were first gated by the bead size (in an FSC/SSC view) and then by the FITC/ATTO-565 – fluorescence ratio giving the well coordinates. The extent of antigen-binding for each POI-SpyCatcher was determined by analyzing the median fluorescence intensity (MFI) of each gated bead population in the Cy5 channel (as shown, right). The wells supplemented with Ø 5 and 10 μm beads were separately analyzed for antigen-binding as the MFI levels are not comparable on different sized particles.