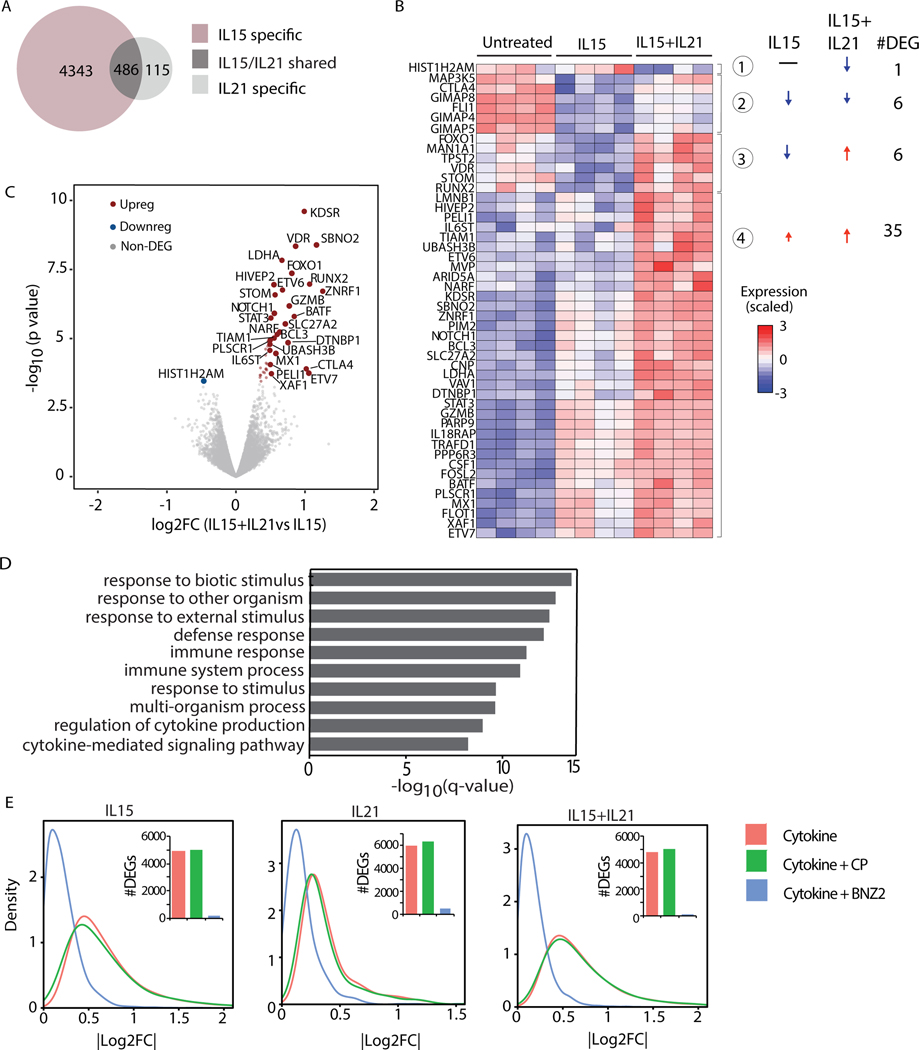
Figure 2. BNZ-2 blocks the transcriptional program individually and cooperatively induced by IL15 and IL21 in tissue-resident IE-CTL.
(A) Venn Diagram displaying the numbers of differentially expressed genes (DEGs) in response to 700pM IL15 or 20pM IL21. The expression of 4343 genes changed in response to IL15 (FDRIL15<0.05 and FDRIL21≥0.05, IL15 specific, pink), while 115 in response to IL21 (FDRIL15≥0.05 and a FDRIL21<0.05, IL21 specific, light grey). 486 genes are changed by both cytokines (FDRIL15<0.05 and FDRIL21<0.05, IL15/IL21 shared, dark grey). (B) Heat-map displaying changes in expression levels upon stimulation with IL15 alone (FDRIL15<0.05) or IL15+IL21 (FDRIL15<0.05 and FDRIL21<0.05) combination compared to untreated IE-CTL lines. Only DEGs in response to IL21+IL15 vs IL15 alone are dispayed: 4 distinct groups were identified based on their change in expression levels. Arrows indicate up- vs down-regulation. Numbers of DEGs in each group are indicated. (C) Volcano plot displaying DEGs in response to the combination of IL15+IL21 vs IL15. Expression levels upon stimulation with IL15 were used as a baseline. p-values are on the y-axis, log2 fold changes (log2FC) in expression upon IL21 addition (IL15+IL21 vs IL15) are on the x axis. The up-regulated genes are in red on the right side, the only downregulated gene is in blue on the left. (D) Gene Ontology (GO) enrichment analysis performed including all up-regulated genes (N=41) when IL21 is added to IL15 (red dots in B), ranked based on p-values. Corrected p-value (q-value) for enrichment of DEGs in each GO term is dispayed. (E) Density plots representing the impact of 1uM BNZ-2 (blue) on IL15 and/or IL21-induced (red) differential gene expression in IE-CTL lines. A scrambled sequence of BNZ-2 served as control peptide (CP, green). Absolute log2FC are on the x axis. Number of DEGs is in the histograms. All data (A-E) are based on gene expression levels in human IE-CTL lines as assessed by RNA-seq. FDR<0.05 was used as a cut-off to evaluate DEGs.