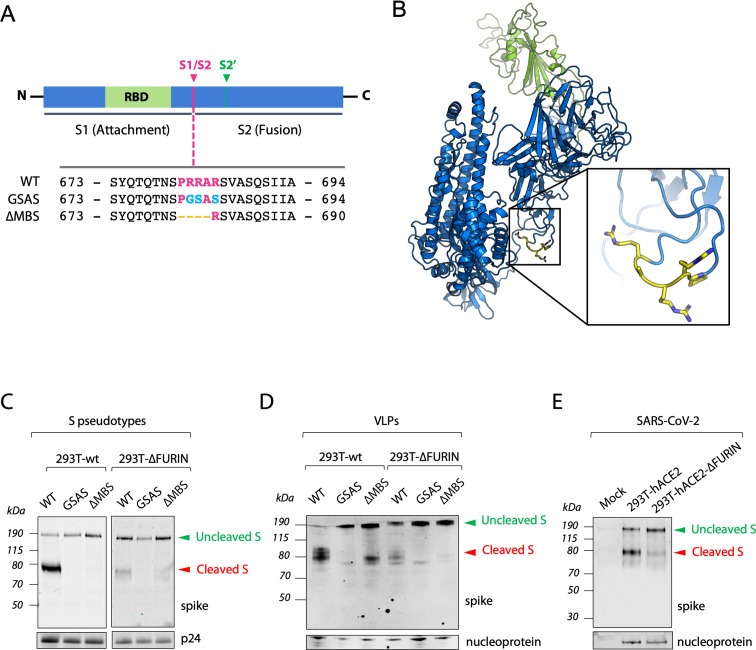
Fig 2. Furin is not essential for the cleavage of S protein but enhances its processing.
(A) Schematic illustration of SARS-CoV-2 S including receptor binding domain (RBD) in green and proteolytic cleavage sites (S1/S2, S2’). Amino acid sequences around the S1/S2 recognition sites of SARS-CoV-2 S are indicated while the multibasic site is highlighted in purple. Amino acid mutations are highlighted in light blue while deletions are marked with orange dashes. (B) Overall structure of the SARS-CoV-2 S protein (PDB: 6VYB). RBD core is shown in green. Pro-Arg-Arg-Ala-Arg residues are shown in yellow. (C,D) Representative western blots of HIV Pseudoviruses (C) and Virus Like Particles (VLPs) (D) harbouring the indicated SARS-CoV-2 S protein mutants (detected with anti-S antibody) and produced in 293-wt and 293T-ΔFURIN cells. Expression of HIV capsid protein (p24) (C) and SARS-CoV-2 nucleoprotein (D) is shown as loading control. (E) Representative western blot analysis of spike and nucleoprotein present in SARS-CoV-2 viral particles produced in 293T-hACE2 and 293T-hACE2-ΔFURIN after 42 hours post infection. The cleaved S in (C) (D) and (E) identifies the S2 subunit.