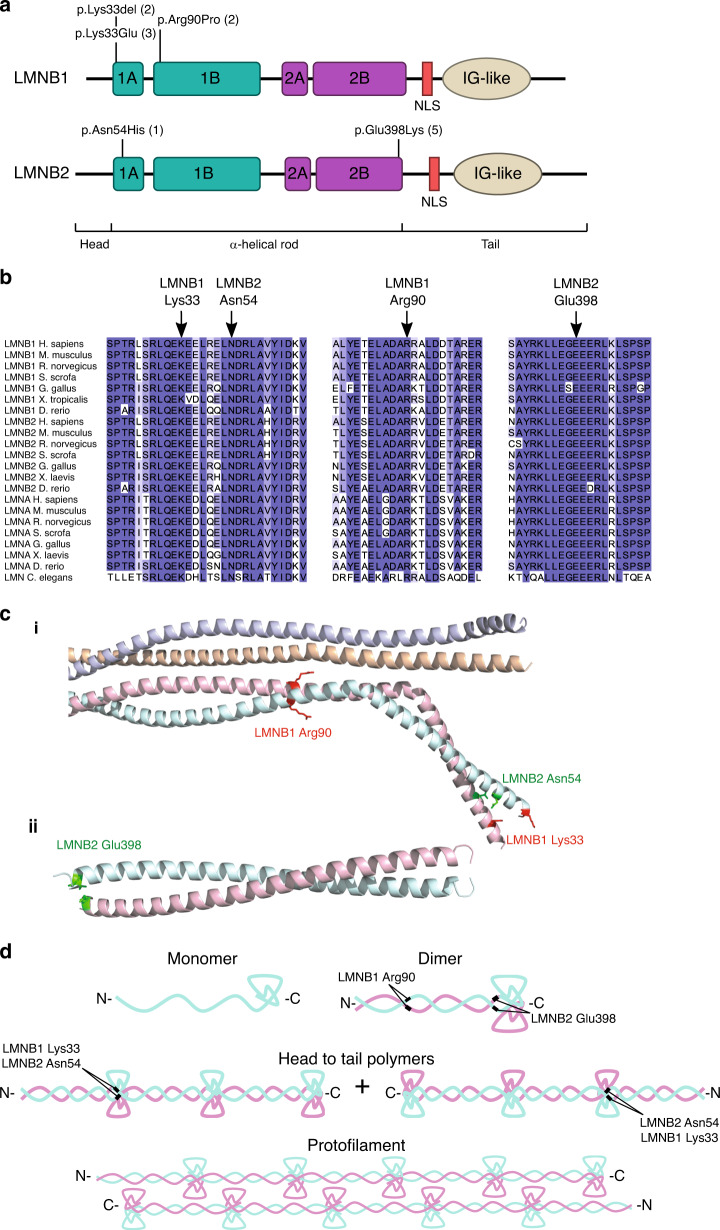
Fig. 1. Pathogenic LMNB1 and LMNB2 variants occur recurrently at highly conserved residues at intra- and interdimer filament interfaces.
(a) Schematic of B-type lamin protein structure. Locations of variants identified in microcephaly patients indicated, number of occurrences in parentheses. The ɑ-helical rod comprises coiled-coil domains 1A, 1B and 2A, 2C, indicated by green and purple boxes respectively. IG-like, immunoglobulin-like domain; NLS, nuclear localization signal. (b) Variants alter residues conserved in all vertebrate and invertebrate lamins. Lamins are specific to metazoa and the number of lamin genes has increased during evolution, with a single lamin gene in C. elegans compared with three mammalian lamin genes. Orthologs of both A- and B-type lamins aligned using Clustal X and colored by percent identity using Jalview. (c) Location of altered residues within lamin structures. While LMNB1/2 structures covering the mutated residues have not been published, as all residues are conserved with LMNA, we could use crystal structures of LMNA with Protein Data Bank (PDB) accessions 6JLB (i) and 1X8Y (ii) for molecular modeling and visualization. The upper structures shows a LMNA homotetramer, with the mutated residues highlighted on one of the constituent homodimers, while the bottom structure is of a LMNA homodimer, as no higher-order structure is available for this region. (d) Lamin filaments are assembled in a hierarchical fashion. First, lamin proteins form dimers, which further assemble into head to tail polymers. These polymers then laterally assemble in an antiparallel fashion to form lamin protofilaments.