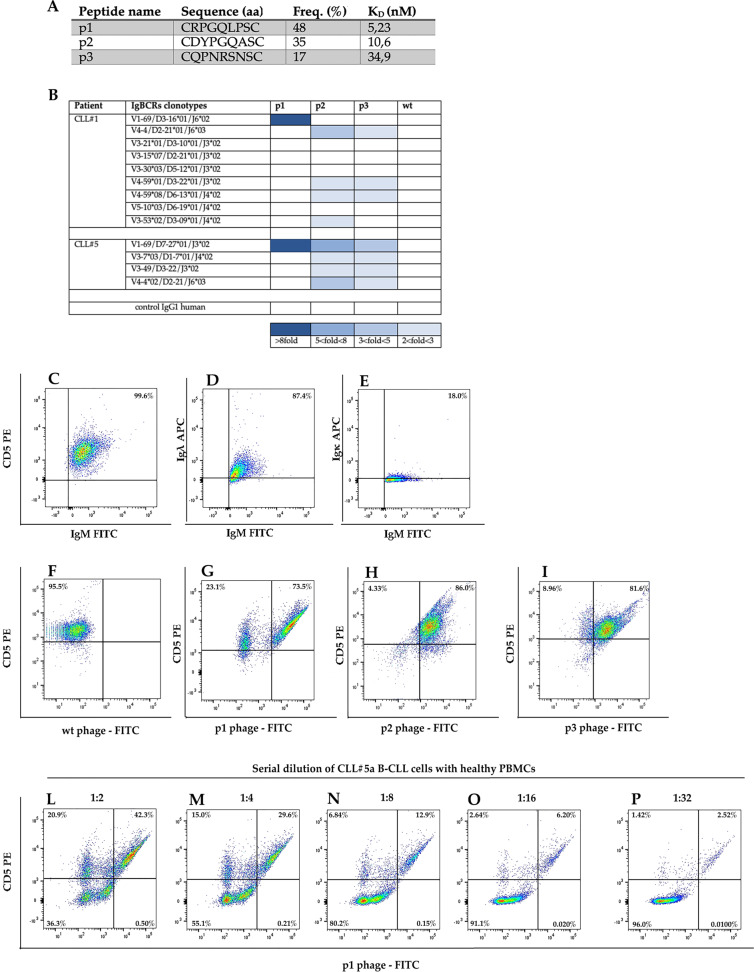
Fig. 1. Analysis of phage ligands of CLL IgBCR in patients CLL#1 and CLL#5.
a Phages ligands of VH1-69 U-CLL IgBCR of CLL#5 patient. RPL screening was performed using the recombinant VH1-69 U-CLL Ig of patient CLL#5 as bait. The phages p1, p2, and p3 were selected at the indicated frequency; DNA sequencing determined the amino acid (aa) sequence of the insert random peptide. The affinity binding was measured as KD by Scatchard Plot analysis of the experiment shown in Supplementary Fig. S4. b Profile of phage binding to the CLL IgBCRs of patient CLL#1 and CLL#5. The recombinant IgBCR (10 ng/μl) was incubated in 96-microwells with the indicated phages (1 × 109 PFU/μl). The phage binding was revealed by the anti-M13 HRP conjugated antibody (Abcam—UK) and relative enzyme substrate. Absorbance was calculated at 405 nm by the MultiskanTM GO Microplate Spectrophotometer (Thermo Fisher Scientific—USA). Wild-type phage and a human IgG were included as controls. Dark blue corresponds to the highest absorbance (>8-fold respect to the blank); decreasing shades of blue correspond to lower absorbance values, as indicated at the bottom of the table. White squares indicate lack of binding. c–p Phage-based flow cytometry of CLL clones. B cells of patient CLL#5 collected at month 1 were analyzed by flow cytometry for the expression of CD5, IgM, and the Igλ and Igκ light chains c–e. The same B-cell sample was analyzed for CD5 expression and phage binding by incubation with the phages wild type (wt) (f), p1 (g), p2 (h), and p3 (i). The B-CLL sample was serially diluted with healthy PBMCs (1:2, 1:4, 1:8, 1:16, 1:32) and analyzed for the binding of phage p1 (l–p). Data were acquired by FACS Canto II (Miltenyi Biotec—Germany) and analyzed by FlowJo Software.