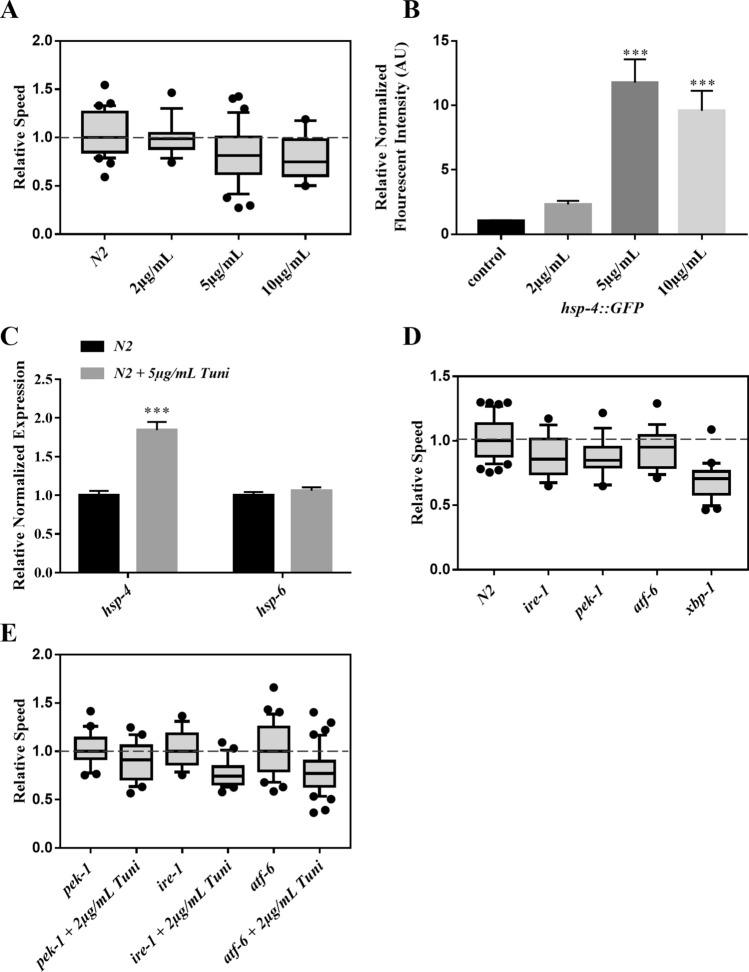
Figure 3.
Electrotaxis phenotype of tunicamycin-treated animals and UPRER mutants. Refer to Fig. 1 for a description of box plot. (A) Wild-type (N2) animals show no speed abnormalities when exposed to 2 μg/mL of tunicamycin (p = 0.590), but exhibit speed deficits at higher doses, 5 μg/mL (p = 0.002) and 10 μg/mL (p = 0.008). (B) Fluorescence intensity of tunicamycin-treated hsp-4::GFP animals. The fluorescence was significantly increased following treatments with 5 μg/mL, and 10 μg/mL tunicamycin (p < 0.001). C) RT-qPCR analysis of chaperons hsp-4 (p = 0.003) and hsp-6 (p = 0.3652) in N2 day-1 adults following exposure to 5 μg/mL of Tunicamycin. (D) Electrotaxis speed of N2 and UPRER mutants. While ire-1(v33) (p = 0.0144), pek-1(ok275) (p = 0.0082) and xbp-1(zc12) (p = 0.0001) animals exhibited significant speed defects, atf-6(ok551) mutants were not significantly affected (p = 0.2995). (E) Responses of mutants following 2 μg/mL tunicamycin treatments., Animals showed a significant decline in the speed. P values are 0.0333 for pek-1(ok275), < 0.0001 for ire-1(v33), and 0.0004 for atf-6(ok551). The numbers of animals were: (A) N2 untreated: n = 30, N2 + 2 μg/mL Tuni: n = 15, N2 + 5 μg/mL Tuni: n = 32, N2 + 10 μg/mL Tuni: n = 10. (B) hsp-4::GFP untreated: n = 45, hsp-4::GFP + 2 μg/mL Tuni : n = 32, hsp-4::GFP + 5 μg/mL Tuni: n = 37, hsp-4::GFP + 10 μg/mL Tuni: n = 31. (C) N2 untreated: n = 3 batches, N2 + 5 μg/mL Tuni: n = 3 batches. (D) N2: n = 42, ire-1: n = 17, pek-1: n = 18, atf-6: n = 18, xbp-1: n = 23. (E) pek-1: n = 18, pek-1 + 2 μg/mL Tuni: n = 20, ire-1: n = 17, ire-1 + 2 μg/mL Tuni: n = 21, atf-6: n = 33, atf-6 + 2 μg/mL Tuni: n = 44. One-way ANOVA was performed followed by Dunnett’s post hoc test for A and B, which showed that all conditions except one (i.e., control vs. 2 µg in panel B, p = 0.7410) were significant. Student’s unpaired t-test was used for the remaining drug conditions. Data in C was analyzed using one-way ANOVA with Tukey’s post hoc test.