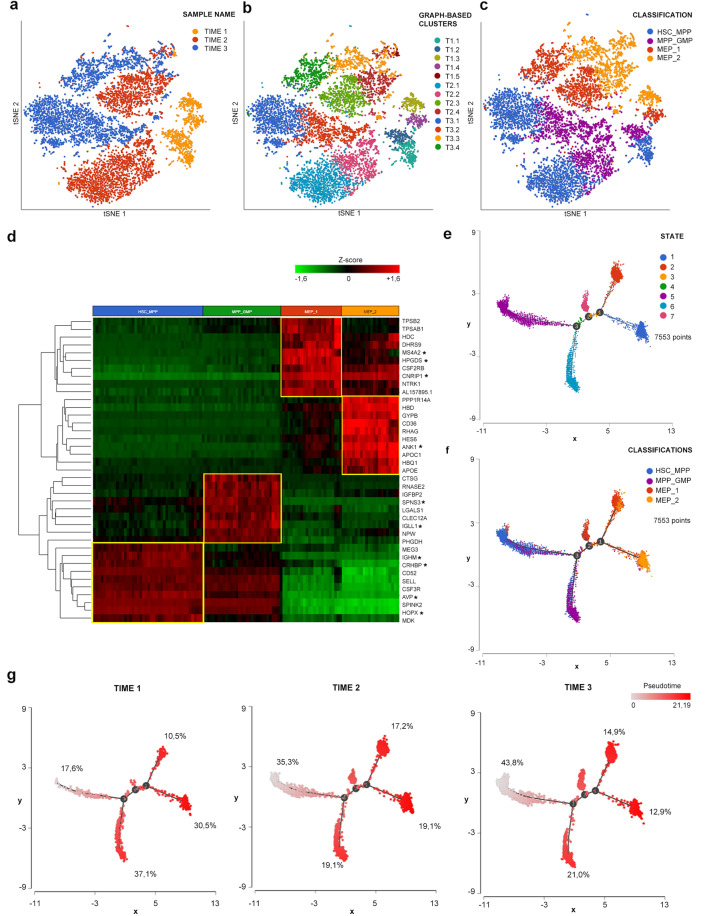
Fig. 3. scRNA-seq clustering and trajectory analysis.
a–c tSNE projections of cells from all three samples in which 7553 cells were included. a Cells are distinguished according to the sample of origin. b Different colors indicate clusters identified by clustering analysis. c The classification we applied to these clusters. We identified four cell groups namely HSC_MPP, MPP_GMP, MEP_1, and MEP_2. d The heatmap was generated using the top 10 marker genes for each cell group. Marker genes shared by the three samples are highlighted by the star next to the gene symbol. e The distribution of single cells colored according to the seven states identified by trajectory analysis. As shown in f, state 5 represented the most primitive one enriched in cells belonging to HSC_MPP cluster. Starting from state 5 the first branch point corresponds to the appearance of a population enriched in MPP_GMP cells. State 7 originates from the second branch point. Two different MEP populations originated from the third branch point, corresponding to MEP_1 and MEP_2 clusters. In g panel trajectories, cells are divided according to the sample of origin. Cells are colored according to pseudotime. Percentages represent the frequency of cells belonging to the indicated sample included in that specific state.