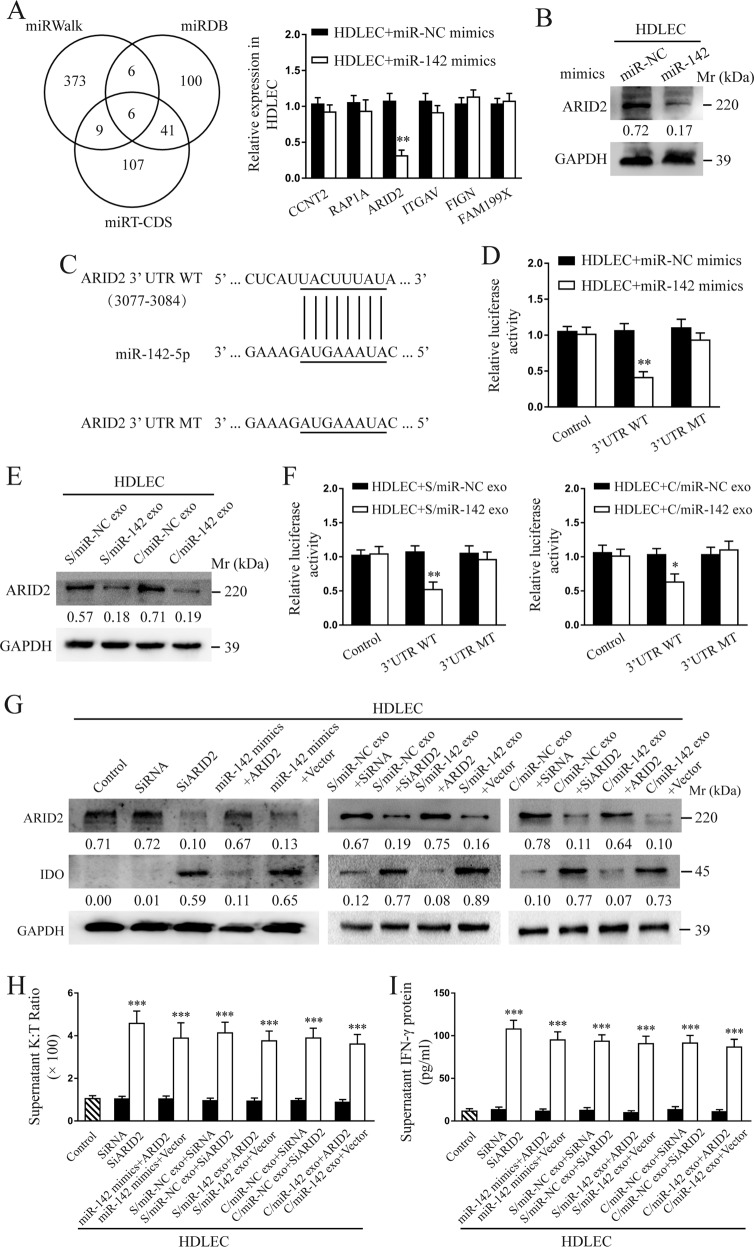
Fig. 4. CSCC-secreted exosomal miR-142-5p directly targets ARID2 in HDLECs.
a Overlap of three miRNA target bioinformatic prediction algorithms. The expression of CCNT2, RAP1A, ARID2, ITGAV, FIGN, and FAM199X in HDLECs pre-treated with miR-142-5p mimics or NC was detected by RT-qPCR. b The protein level of ARID2 in HDLECs pre-treated with miR-142-5p mimics or NC was analysed by western blotting. c The wild-type- and mutated-binding sites between miR-142-5p and ARID2 were shown. d Relative luciferase activity of HDLECs pre-treated with miR-142-5p mimics or NC was analysed. e The protein level of ARID2 in HDLECs pre-treated with indicated exosomes was analysed by western blotting. f Relative luciferase activity of HDLECs pre-treated with indicated exosomes was analysed. g The expression of ARID2 and IDO proteins in miR-142-5p-mimic/exosome-treated HDLECs transfected with plasmid-ARID2 or vector control and SiARID2- or SiRNA-treated HDLECs was detected by western blotting. h The K:T ratio of HDLEC supernatant after the indicated treatment was calculated. i IFN-γ protein level in the supernatant of HDLECs following indicated treatment was detected by ELISA. miR-142, miR-142-5p. S/miR-142 Siha/miR-142-5p, C/miR-NC Caski/miR-NC, C/miR-142 Caski/miR-142-5p, exo exosomes, WT wild-type, MT mutant. Error bars represent the mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.