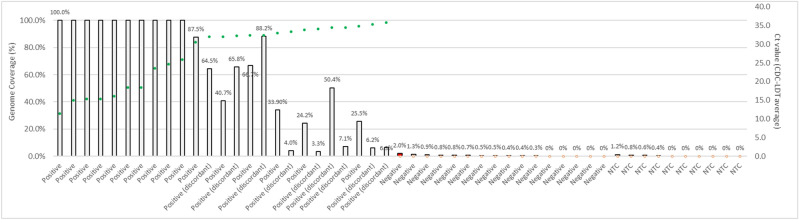
Figure 3.
Correlation between viral genome coverage (%) by next generation sequencing (NGS) and cycle threshold (Ct) values for SARS CoV-2. NGS is a sensitive method of detecting low-level SARS-CoV-2 virus in clinical samples, and coverage (%) of the genome is correlated to the Ct value determined by RT-PCR. Thirty samples (17 saliva and 11 SN swabs and 1 NP swab) were tested by NGS. Ten samples (4 saliva, 6 SN swabs) with discordant results from the RT-PCR tests were confirmed to be positive by NGS (median genome coverage 29.7%, range 3.3–88.2%). Genome coverage (%) is defined as the proportion of the SARS-CoV-2 genome that has > 1× depth of coverage. The threshold coverage (%) for making positive call by NGS was established by running 5 negatives (by RT-PCR) samples from this study, 11 negative NP swab samples from community testing, and 10 no-template controls (NTC). The threshold was determined to be the detection of 5 amplicons, which corresponds to a coverage of ~ 1.7% of the SARS-CoV-2 genome. Among five negatives (by both RT-PCR methods) saliva samples, one saliva sample was called positive by NGS (red bar) with a genome coverage of 2.0%. This sample was from a patient with RT-PCR positive NP swab and nasal swab samples collected at the same time, raising the possibility of a low-level signal detected on NGS. Ct values for samples with positive calls (by either or both Fortitude 2.1 and CDC-LDT RT-PCR assays) are represented on the graph with green circles, and samples negative for SARS-CoV-2 (by both RT-PCR tests) and NTC are represented as open circles. Average Ct values (of 2 targets) or single Ct value (when only 1 target was detected) from the CDC-LDT assay are plotted on the secondary axis.