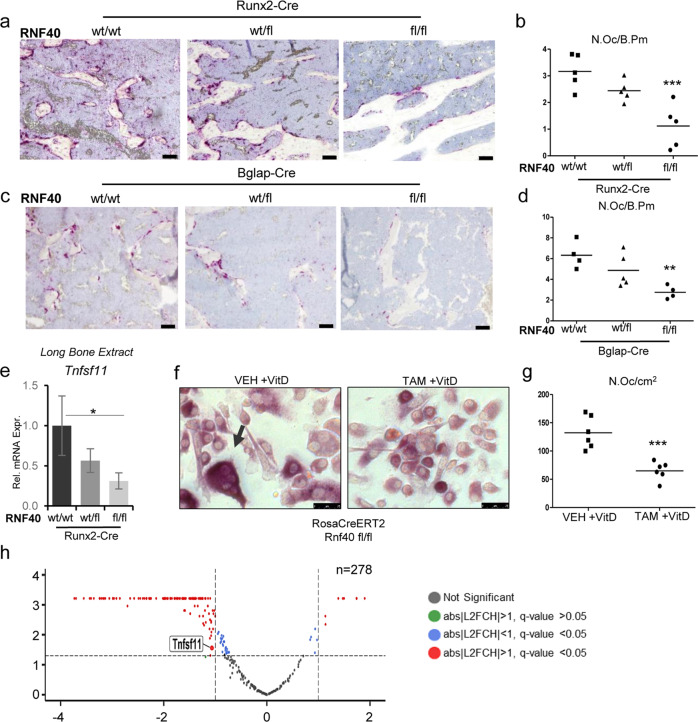
Fig. 4. RNF40 is a negative regulator of bone mass.
The plastic-embedded femoral sections from a Rnf40Runx2-Cre and c Rnf40Bglap-Cre were subjected to TRAP staining. Bar: 50 µm. b–d The number of osteoclasts was defined using “osteomeasure” software and depicted as number of osteoclasts per bone perimeter (N.Oc/B.PM) for each respective genotype. e The relative expression of Rankl (Tnfsf11) was evaluated in the tibae of Rnf40Runx2-Cre wild-type (wt/wt), heterozygous (wt/fl), and knockout (fl/fl) mice via qRT-PCR analysis and normalized to the expression level of the Gapdh housekeeping gene. Mean ± SD, n = 3. One-way ANOVA followed by Bonferroni’s test was performed for statistical analysis where ***p < 0.001, **p < 0.01, *p < 0.05. f, g Primary calvarial osteoblasts from Rnf40 mouse with an initial 7 day TAM or VEH treatment were cocultured with primary bone marrow-derived macrophages from wild-type mice for 9 days with the addition of 10 nM vitamin D treatment. TRAP staining was used to visualize the osteoclasts (f). Quantification of TRAP-positive multinucleated osteoclasts from the coculture experiment defined in g. h Primary calvarial osteoblasts from Rnf40Rosa-CreERT2 subjected to 7 days of 4-hydroxytamoxifen (TAM) or vehicle (VEH) treatment were subsequently treated with vitamin D (+vitD) or an equivalent volume of DMSO (−vitD) for an additional 7 days. RNA was harvested and subjected to sequencing. The volcano plot shows vitamin D-induced genes (n = 278, q value ≤ 0.05, log2 fold change (L2FC) ≥ 0.8) and their regulation by Rnf40 knockout. Red color indicates significantly up- or downregulated genes in the Rnf40 knockout with abs|L2FC| ≥ 1 and q value ≤ 0.05; the blue dots—significantly (q value ≤ 0.05) regulated genes with less than abs|L2FC| < 1; the green dots—insignificantly regulated genes with abs|L2FC| ≥ 1 and q value > 0.05; and the black dots—unregulated genes with q value > 0.05 and abs|L2FC| < 1.