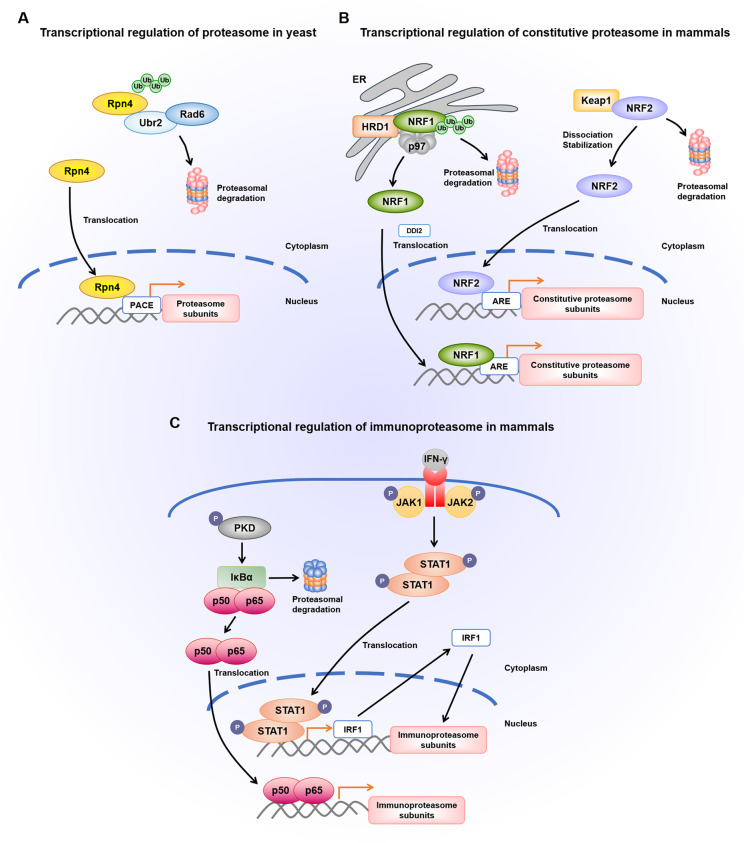
Fig. 2. Transcriptional regulation of proteasome expression.
A Transcriptional regulation of proteasome in yeast. Rpn4 serves as a transcription factor with short half-life (t1/2 ~2 min) owing to the proteasomal degradation mediated by E2 ubiquitin conjugating enzyme Rad6 and E3 ubiquitin ligase Ubr2. Upon proteasome inhibition, Rpn4 is translocated into the nucleus, where it binds to PACE sequence in the promoters of proteasome subunit genes, resulting in the compensatory increase of proteasome expression. B Transcriptional regulation of constitutive proteasome in mammals. NRF1 resides in the ER, which is degraded via ERAD pathway requiring ER-resident ubiquitin ligase HRD1 and ATPase p97. When the proteasome is inhibited, NRF1 is cleaved by DDI2, and then translocated to the nucleus where it binds ARE and activates the transcription of proteasome genes. During oxidative stress, NRF2-Keap1 complex disassociates, and NRF2 translocates into the nucleus to transcriptionally regulate proteasome expression. C Transcriptional regulation of immunoproteasome in mammals. Under the stimulation of IFN-γ, activated JAK1 and JAK2 leads to the dimerization and phosphorylation of STAT1, which translocates into the nucleus and binds to IRF1. Following translation, IRF1 moves back into the nucleus to increase the transcription of immunoproteasome. Upon oxidative injury, phosphorylated protein kinase D (PKD) leads to the disassociation of IκBα from NF-κB. In turn, IκBα is degraded and NF-κB translocates into the nucleus to regulate the expression of immunoproteasome.